 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009602857.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 753aa MW: 82825 Da PI: 5.9799 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66 | 5.2e-21 | 55 | 110 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l+L++rqVk+WFqNrR+++k
XP_009602857.1 55 KKRYHRHTPQQIQELESLFKECPHPDEKQRLELSKRLSLETRQVKFWFQNRRTQMK 110
688999***********************************************999 PP
| |||||||
| 2 | START | 202.4 | 1.9e-63 | 262 | 486 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetl 82
ela++a++e+vk+a+ +ep+W++s+ e++n +e++++f++ + + +ea+r++g+v+ ++ lve+l+d++ +W e+++ + +t+
XP_009602857.1 262 ELALAAMDEFVKMAQTDEPLWFRSVeggrEILNHEEYMRSFTSCIGmrpnsFVSEASRETGMVIINSLALVETLMDSN-KWAEMFPcliaRTSTT 355
5899*************************************988889***9***************************.**************** PP
EEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-E CS
START 83 evissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehv 170
+vissg galqlm aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+d ++ ++ + + ++ lpSg+l+++++ng+skvtwveh
XP_009602857.1 356 DVISSGmggtrnGALQLMHAELQVLSPLVPiREVNFLRFCKQHAEGVWAVVDVSIDTIRETSNAPTFPNSRILPSGCLVQDMPNGYSKVTWVEHG 450
*************************************************************99******************************** PP
E--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 171 dlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+++++++h l+r+l++ g+ +ga++wvatlqrqce+
XP_009602857.1 451 EYDESVIHHLYRPLISAGMGFGAQRWVATLQRQCEC 486
**********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.3E-20 | 41 | 112 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-21 | 41 | 106 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.637 | 52 | 112 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.2E-19 | 53 | 116 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.74E-19 | 54 | 112 | No hit | No description |
| Pfam | PF00046 | 1.5E-18 | 55 | 110 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 87 | 110 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 42.638 | 253 | 489 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.47E-32 | 255 | 486 | No hit | No description |
| CDD | cd08875 | 1.24E-124 | 257 | 485 | No hit | No description |
| Pfam | PF01852 | 4.5E-55 | 262 | 486 | IPR002913 | START domain |
| SMART | SM00234 | 5.0E-48 | 262 | 486 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.83E-23 | 505 | 739 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 753 aa Download sequence Send to blast |
MEGQSEVTRM AENYEGNNSN GRRSREEEPD SRSGSDNLEG ASGDDQDAAD KPPRKKRYHR 60 HTPQQIQELE SLFKECPHPD EKQRLELSKR LSLETRQVKF WFQNRRTQMK TQLERHENSI 120 LRQENDKLRA ENMSIREAMR NPICTNCGGP AMIGEISLEE QHLRIENARL KDELDRVCAL 180 AGKFLGRPIS SLVNSMPPPM PNSSLELGVG NNGFGGLSNV PSTLPLAPPD FGVGISNSLP 240 VVASTRQTTG IERSLERSMY LELALAAMDE FVKMAQTDEP LWFRSVEGGR EILNHEEYMR 300 SFTSCIGMRP NSFVSEASRE TGMVIINSLA LVETLMDSNK WAEMFPCLIA RTSTTDVISS 360 GMGGTRNGAL QLMHAELQVL SPLVPIREVN FLRFCKQHAE GVWAVVDVSI DTIRETSNAP 420 TFPNSRILPS GCLVQDMPNG YSKVTWVEHG EYDESVIHHL YRPLISAGMG FGAQRWVATL 480 QRQCECLAIL MSSTVSSRDH TAITPSGRRS MLKLAQRMTN NFCAGVCAST VHKWNKLCAG 540 NVDEDVRVMT RKSVDDPGEP PGIVLSAATS VWLPISPQRL FDFLRDERLR SEWDILSNGG 600 PMQEMAHIAK GQDHGNCVSL LRASAINANQ SSMLILQETC IDAAGALVVY APVDIPAMHV 660 VMNGGDSAYV ALLPSGFSIV PDGPGSRGSS TANCGSNNDG PDQRISGSLL TVAFQILVNS 720 LPTAKLTVES VETVNNLISC TVQKIKAALH CES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
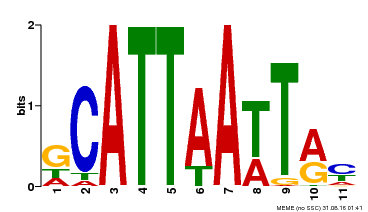
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GQ222185 | 0.0 | GQ222185.1 Solanum lycopersicum cultivar M82 cutin deficient 2 (CD2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009602856.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2-like isoform X1 | ||||
| Refseq | XP_016490208.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2-like | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A1S4BMT8 | 0.0 | A0A1S4BMT8_TOBAC; homeobox-leucine zipper protein ANTHOCYANINLESS 2-like | ||||
| STRING | XP_009602856.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



