 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009620290.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 766aa MW: 83555.7 Da PI: 6.6747 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.5 | 4.6e-16 | 24 | 68 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE+ ++++a k++G W +I +++g ++t+ q++s+ qk+
XP_009620290.1 24 RERWTEEEHNRFLEALKLYGRA-WQRIEEHIG-TKTAVQIRSHAQKF 68
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 8.22E-17 | 18 | 74 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.173 | 19 | 73 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 7.0E-17 | 22 | 71 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.1E-12 | 23 | 71 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-8 | 24 | 64 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.2E-13 | 24 | 67 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.49E-9 | 26 | 69 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0042754 | Biological Process | negative regulation of circadian rhythm | ||||
| GO:0043433 | Biological Process | negative regulation of sequence-specific DNA binding transcription factor activity | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 766 aa Download sequence Send to blast |
MDSYSSGEEL VVKTRKPYTI TKQRERWTEE EHNRFLEALK LYGRAWQRIE EHIGTKTAVQ 60 IRSHAQKFFT KLEKEAVIKG VPISQALDIE IPPPRPKRKP SNPYPRKTSV AAPSSQVGIK 120 DGKLSPFSSI CEDRNLFDLE KEPITEKPGR NEKLGSVQEN QNKKNCSQGF ALFKEGASAP 180 SLSPGKSLQT LAEPAGSCTL NESLPATKGV INHDVTAKSF LTVGSKEHQK LNILDAKQSF 240 QSNSSCNTFN EGKSCQSNEK LAQDEKKDQP SQPDHFGKFS RNDMQVPHHY PRHVPVHILD 300 GALGVSGAQT TPDMFYPESV SHQIGGVQGL SNLYTNPTSS ATSEHHSNAA MSSIHQSFPC 360 FHPNFTPIHD PDDYRSFLQL SSTFSSLIFS ALLQNPAAHA AASFAASFWP YANMEAPVDS 420 PTGNTASQIN SAPSMAAIAT ATVAAATAWW AAHGLLPLCS PFHSSSTRVP TSATSMQVDP 480 CQPSVGKNEG REGSHNSPHA QQTVPDCSEA LQEHHSASEL PTSPSSESEE SEGRKLKTGL 540 TANDTEQGAA VTEIHEPNTG NGRKQVDRSS CGSNTPSSSD LETDALEKDE KGKEEPQEPN 600 VNLLAGDAGN RRGRNCISPN DFWKEVSEGG RIAFQALFTR EKLPQSFSPS NDLKNKGKIN 660 LENVKEKPDE KGLSGSQLDL NDQASDMCSS HQAVEDNVLV IGNKEDAEKC LPIMELGQVR 720 LKARRTGFKP YKRCSLEAKD SRVTSSSCQD EEKSAKRLRL EGEASI |
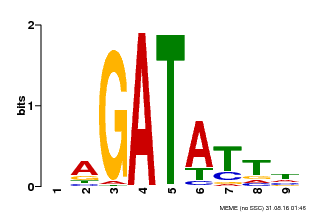
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00654 | PBM | Transfer from LOC_Os08g06110 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JQ424913 | 0.0 | JQ424913.1 Nicotiana attenuata late elongated hypocotyl (LHY) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009620290.1 | 0.0 | PREDICTED: protein LHY | ||||
| Refseq | XP_018631810.1 | 0.0 | PREDICTED: protein LHY | ||||
| Refseq | XP_018631811.1 | 0.0 | PREDICTED: protein LHY | ||||
| TrEMBL | A0A1S3ZFF1 | 0.0 | A0A1S3ZFF1_TOBAC; protein LHY-like | ||||
| STRING | XP_009620290.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA9710 | 13 | 15 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01060.4 | 8e-49 | MYB_related family protein | ||||



