 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009757585.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 555aa MW: 60351.7 Da PI: 8.1433 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.7 | 1.6e-16 | 259 | 308 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lr ++P++ ++K +Ka+ L +++eYI+ Lq
XP_009757585.1 259 RSKHSATEQRRRSKINDRFQMLRGIIPNS----DQKRDKASFLLEVIEYIHFLQ 308
889*************************9....9******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.1E-23 | 254 | 325 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.07E-18 | 255 | 325 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.272 | 257 | 307 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.8E-14 | 259 | 308 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.01E-15 | 259 | 312 | No hit | No description |
| SMART | SM00353 | 8.4E-13 | 263 | 313 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 555 aa Download sequence Send to blast |
MELPQPRPFG TEGRKTTHDF LSLYSPVQQD PTPPQGGFLK THDFLQPLEQ AEKALRKEEA 60 KVEVVAVEKP PAPVASPSVE HILPGGIGTY SISYLHQRIP KPEASLFAVA QATSTDRNDE 120 NSNCSSYTGG SGFTLWDESA VKGKTGKENS GGDRHALREA GLNTGGGQPT TSLEWQSQSS 180 SNHKHNTTAI SSLSSARQSS PLKTQSFVHM ITSAKNAQDD DDDDDEDFVI KKEPQSHPRG 240 NLSVKVDGEG NDKKPNTPRS KHSATEQRRR SKINDRFQML RGIIPNSDQK RDKASFLLEV 300 IEYIHFLQEK VHKYEESYQG WENEPLKLPL SKCHRTTQGA SNHPQGTINA SGAAPTYAVK 360 FDENIMGISS TNPFNAQKVE PNISTTSLKE SSQRPGLTNK ATTPSMSPNT FPFCGASTAA 420 LYSSKLTADT AKLESKSHPQ FSVSRSHMTD YAIPNANSKG QDLSIESGTI SISSAYSQGL 480 LNTLTLALQN SGVDLSQANI SVQIDLGKRA NGRLHSSAST VKGDDVSTSN QPIPKSISTS 540 TREESDRAFK RLKTS |
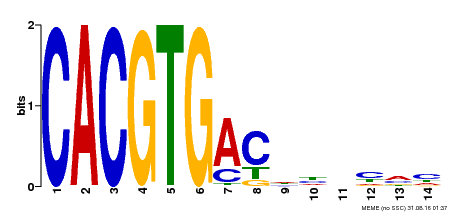
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT013786 | 0.0 | BT013786.1 Lycopersicon esculentum clone 132679F, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009757585.1 | 0.0 | PREDICTED: transcription factor BIM1 isoform X2 | ||||
| Refseq | XP_016514052.1 | 0.0 | PREDICTED: transcription factor BIM1-like isoform X2 | ||||
| TrEMBL | A0A1S4DKZ2 | 0.0 | A0A1S4DKZ2_TOBAC; transcription factor BIM1-like isoform X2 | ||||
| TrEMBL | A0A1U7V764 | 0.0 | A0A1U7V764_NICSY; transcription factor BIM1 isoform X2 | ||||
| STRING | XP_009757584.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 1e-108 | bHLH family protein | ||||



