 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009757586.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 554aa MW: 60222.6 Da PI: 8.4352 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.7 | 1.6e-16 | 258 | 307 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lr ++P++ ++K +Ka+ L +++eYI+ Lq
XP_009757586.1 258 RSKHSATEQRRRSKINDRFQMLRGIIPNS----DQKRDKASFLLEVIEYIHFLQ 307
889*************************9....9******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.1E-23 | 253 | 324 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.07E-18 | 254 | 324 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.272 | 256 | 306 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.92E-15 | 258 | 311 | No hit | No description |
| Pfam | PF00010 | 3.8E-14 | 258 | 307 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.4E-13 | 262 | 312 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 554 aa Download sequence Send to blast |
MELPQPRPFG TEGRKTTHDF LSLYSPVQQD PTPPQAGGFL KTHDFLQPLE QAEKALRKEE 60 AKVEVVAVEK PPAPVASPSV EHILPGGIGT YSISYLHQRI PKPEASLFAV AQATSTDRND 120 ENSNCSSYTG GSGFTLWDES AVKGKTGKEN SGGDRHALRG LNTGGGQPTT SLEWQSQSSS 180 NHKHNTTAIS SLSSARQSSP LKTQSFVHMI TSAKNAQDDD DDDDEDFVIK KEPQSHPRGN 240 LSVKVDGEGN DKKPNTPRSK HSATEQRRRS KINDRFQMLR GIIPNSDQKR DKASFLLEVI 300 EYIHFLQEKV HKYEESYQGW ENEPLKLPLS KCHRTTQGAS NHPQGTINAS GAAPTYAVKF 360 DENIMGISST NPFNAQKVEP NISTTSLKES SQRPGLTNKA TTPSMSPNTF PFCGASTAAL 420 YSSKLTADTA KLESKSHPQF SVSRSHMTDY AIPNANSKGQ DLSIESGTIS ISSAYSQGLL 480 NTLTLALQNS GVDLSQANIS VQIDLGKRAN GRLHSSASTV KGDDVSTSNQ PIPKSISTST 540 REESDRAFKR LKTS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
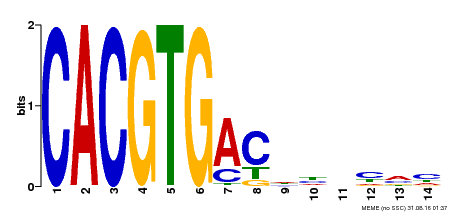
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT013786 | 0.0 | BT013786.1 Lycopersicon esculentum clone 132679F, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009757586.1 | 0.0 | PREDICTED: transcription factor BIM1 isoform X3 | ||||
| Refseq | XP_016514053.1 | 0.0 | PREDICTED: transcription factor BIM1-like isoform X3 | ||||
| TrEMBL | A0A1S4DKY9 | 0.0 | A0A1S4DKY9_TOBAC; transcription factor BIM1-like isoform X3 | ||||
| TrEMBL | A0A1U7UXW3 | 0.0 | A0A1U7UXW3_NICSY; transcription factor BIM1 isoform X3 | ||||
| STRING | XP_009757584.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 1e-106 | bHLH family protein | ||||



