 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009763883.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1102aa MW: 123588 Da PI: 6.0816 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 182.4 | 5.2e-57 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
l+ ++rwl++ ei++iL+n++k++++ e+++rp+sgsl+L++rk++ryfrkDG+sw+kkkdgktv+E+he+LK g+++vl+cyYah+een++fqr
XP_009763883.1 20 LEAQHRWLRPAEICEILKNYQKFRIAPEPPNRPPSGSLFLFDRKVLRYFRKDGHSWRKKKDGKTVKEAHERLKAGSIDVLHCYYAHGEENENFQR 114
5679******************************************************************************************* PP
CG-1 97 rcywlLeeelekivlvhylevk 118
r+yw+Leee+++ivlvhy+evk
XP_009763883.1 115 RSYWMLEEEMSHIVLVHYREVK 136
*******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 83.558 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.9E-79 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.7E-50 | 21 | 134 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 5.3E-4 | 516 | 599 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 9.57E-16 | 517 | 601 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 4.6E-4 | 517 | 593 | IPR002909 | IPT domain |
| CDD | cd00204 | 4.75E-12 | 699 | 808 | No hit | No description |
| PROSITE profile | PS50297 | 17.582 | 699 | 821 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.8E-17 | 700 | 813 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.48E-16 | 704 | 811 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 6.8E-8 | 721 | 812 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.576 | 749 | 782 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1.9 | 749 | 779 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 120 | 789 | 818 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.45E-7 | 918 | 975 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 50 | 924 | 946 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.346 | 926 | 954 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.18 | 927 | 945 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0042 | 947 | 969 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.285 | 948 | 972 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 5.0E-4 | 950 | 969 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1102 aa Download sequence Send to blast |
MADSRRYGLN AQLDIDQILL EAQHRWLRPA EICEILKNYQ KFRIAPEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHSWRKKKD GKTVKEAHER LKAGSIDVLH CYYAHGEENE NFQRRSYWML 120 EEEMSHIVLV HYREVKGNRT NFSRTREPQE ATPRFQETDE DVHSSEVDSS ASTKFYPNGY 180 QVNSQVTDAT SLSSAQASEY EDAESAYNQH PTSGFHSFLD AQPSMMQKAG ESLPVPYHPI 240 PFSNDHQVQF AGSSDMDFFS SAPGNKSRNT ANTYIPSRNL DFPSWETISV NNPAAYQSYH 300 FQPSSQSGAN NMTHEQGSTT MGQVFLNDFK KQGQNRIDSL GDWQTSEGDA AFISKWSMDQ 360 KLNPNLASDH TIRSSAAYNV ELHNSLEASH ILPSHQDKHP MQNELPSQLS DANVGGSLNA 420 ELDHNLSIGV RTDHSSLKQP LLDGVLREGL KKLDSFDRWM SKELEDVSEP HMQSNSSSYW 480 DNVGDDDGVD NSTIASQVQL DTYMLSPSLS QDQFFSIIDF SPSWAFAGSE IKVLITGKFL 540 KSQPEVEKWA CMFGELEVPA EVIADGVLRC HTPNQKVGRV PFYITCSNRL ACSEVREFEF 600 RVSESQDVDV ANSCSSSESL LHMRFGKLLS LESTVSLSSP PRSEDDVSNV CSKINSLLKE 660 DDNEWEEMLN LTYENNFMAE KVKDQLLQKL LKEKLRVWLL QKVAEGGKGP NVLDEGGQGV 720 LHFAAALGYD WAIPPTIAAG VSVNFRDVNG WTALHWAASY GRERTVGFLI ISLGAAPGAL 780 TDPTPKHPSG RTPADLASSN GHKGIAGYLA ESSLSSHLSS LELKEMKQGE TVQPFGEAVQ 840 TVSERSATPA WDGDWPHGVS LKDSLAAVRN ATQAAARIHQ VFRVQSFQRK QLKEHGGSEF 900 GLSDEHALSL LALKTNKAGQ HDEPVHTAAV RIQNKFRSWK GRRDYLLIRQ RIIKIQAHVR 960 GHQVRNKYKN IIWSVGILEK VILRWRRKGS GLRGFKPEAT LTEGSNMQDR PVQEDDYDFL 1020 KEGRKQTEQR LQKALARVKS MVQYPEARDQ YRRLLNVVSD MKDTTTTSDG APSNSGEAAD 1080 FGDDLIDLDD LLDDDTFMST AP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
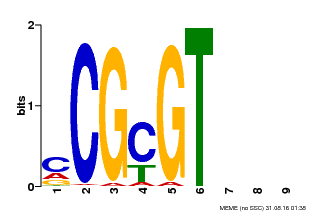
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF253511 | 0.0 | AF253511.1 Nicotiana tabacum anther ethylene-upregulated protein ER1 (ER1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009763883.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3-like isoform X2 | ||||
| Refseq | XP_016478077.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3-like isoform X2 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A1S4AMX5 | 0.0 | A0A1S4AMX5_TOBAC; calmodulin-binding transcription activator 3-like isoform X2 | ||||
| TrEMBL | A0A1U7VBZ1 | 0.0 | A0A1U7VBZ1_NICSY; calmodulin-binding transcription activator 3-like isoform X2 | ||||
| STRING | XP_009763882.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



