 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009776015.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 217aa MW: 23826.6 Da PI: 8.5045 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 22.4 | 2.1e-07 | 141 | 172 | 4 | 39 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-ST CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKls 39
+h+ +Er RR++i +++ L++l+P + +kKl
XP_009776015.1 141 SHSLAERARREKISERMKILQDLVPGC----NKKLH 172
8************************99....67765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 9.61E-6 | 135 | 171 | No hit | No description |
| SuperFamily | SSF47459 | 3.79E-9 | 135 | 173 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 11.439 | 137 | 187 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-9 | 137 | 176 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.8E-5 | 141 | 172 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048446 | Biological Process | petal morphogenesis | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 217 aa Download sequence Send to blast |
MDPMMNEARA FQAAAPNPPF NLAEIWPFPI NAGAAMPYAN GDHHFPINDP MLLDHRTNHR 60 SGSARKRRED DESAKGVSTS GNGLSESDSK RLKATGSNEN HESGGDGEGN SGKSADQPAK 120 SAEPPKDYIH VRARRGQATD SHSLAERARR EKISERMKIL QDLVPGCNKK LHYSQIGSML 180 VSSVDVSITS RAPMQAQPWN LPFLFVITIS FQFVIFS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the control of petal size, by interfering with postmitotic cell expansion to limit final petal cell size. {ECO:0000269|PubMed:16902407}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00207 | DAP | Transfer from AT1G59640 | Download |
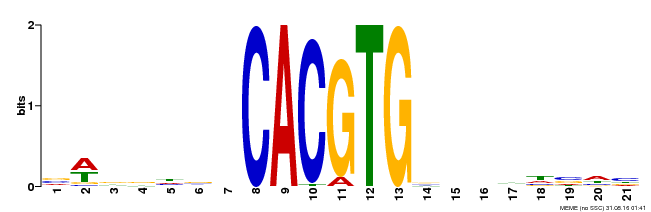
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Isoform 1 is up-regulated by PI/AP3, SEP2, SEP3 and AP1, but repressed by AG. {ECO:0000269|PubMed:16902407}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009776015.1 | 1e-162 | PREDICTED: transcription factor bHLH79 isoform X4 | ||||
| Swissprot | Q0JXE7 | 4e-38 | BPE_ARATH; Transcription factor BPE | ||||
| TrEMBL | A0A1U7WP40 | 1e-160 | A0A1U7WP40_NICSY; transcription factor bHLH79 isoform X4 | ||||
| STRING | XP_009776012.1 | 1e-122 | (Nicotiana sylvestris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59640.2 | 2e-34 | BIG PETAL P | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



