 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009788577.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 661aa MW: 74416.5 Da PI: 6.3398 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 431.8 | 6.5e-132 | 40 | 399 | 1 | 348 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWW 95
eel++rmwkd+++lkrlker+k +++ a++++k++++++qarrkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gasd++raWW
XP_009788577.1 40 EELERRMWKDRVKLKRLKERQKLA--AQQ-AADKQKNKQTTDQARRKKMSRAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGASDNIRAWW 131
8*******************9963..355.59*************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT- CS
EIN3 96 kekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdq 190
kekv+fd+ngpaai+ky+a++ + +++ ++ ++ + ++ l++lqD+tlgSLLs+lmqhcdppqr+fplekgv+pPWWPtG+e+ww+++gl+k q
XP_009788577.1 132 KEKVKFDKNGPAAIAKYEAECRAKGEGVGN--QNGNPQSVLQDLQDATLGSLLSSLMQHCDPPQRKFPLEKGVSPPWWPTGNEEWWAKMGLPKGQ 224
**********************99988888..57999999******************************************************* PP
-.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX..XX.......XXXXXXXXXXX CS
EIN3 191 gtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah..ss.......slrkqspkvtl 276
+ ppykkphdlkk++kv+vLtavikhmsp+i++ir+l+rqsk+lqdkm+akes ++l+vl++ee+ +++ s++ ss +r +++k +
XP_009788577.1 225 K-PPYKKPHDLKKMYKVGVLTAVIKHMSPDIAKIRRLVRQSKCLQDKMTAKESSIWLAVLSREESTIQQPSSDngSSsiseapnRIRGEKKKPSV 318
*.9*********************************************************************97633577777777779999999 PP
XXXXXXXXXXXXXXX.XXXXXXXXXX............XXXXXXXXX.XXXXXXXXXXXX......XXXXXXX.XXXXXXXXXXX CS
EIN3 277 sceqkedvegkkeskikhvqavktta............gfpvvrkrk.kkpsesakvsskevsrtcqssqfrgsetelifadkns 348
++e+++dv+g ++ + + ++ +++ + +++ + +r+ +k+++s+ + +++ ++q ++ e +++++d n+
XP_009788577.1 319 DSESDYDVDGVNDGNDSVSSRDERGNepsnarpqshqgKEQGNGQRRkRKRARSN---P-QQTQLSLNNQQPDDEARNTLPDINN 399
*********5555553444444443355566666665533333333302222222...2.2357889999999999999999776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 2.9E-125 | 40 | 287 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 2.7E-70 | 164 | 295 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.31E-60 | 168 | 290 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 661 aa Download sequence Send to blast |
MEYCMIDADG LGNSSDIEVD DIRCDNIEEK DVSDEEIDPE ELERRMWKDR VKLKRLKERQ 60 KLAAQQAADK QKNKQTTDQA RRKKMSRAQD GILKYMLKLM EVCKARGFVY GIIPEKGKPV 120 SGASDNIRAW WKEKVKFDKN GPAAIAKYEA ECRAKGEGVG NQNGNPQSVL QDLQDATLGS 180 LLSSLMQHCD PPQRKFPLEK GVSPPWWPTG NEEWWAKMGL PKGQKPPYKK PHDLKKMYKV 240 GVLTAVIKHM SPDIAKIRRL VRQSKCLQDK MTAKESSIWL AVLSREESTI QQPSSDNGSS 300 SISEAPNRIR GEKKKPSVDS ESDYDVDGVN DGNDSVSSRD ERGNEPSNAR PQSHQGKEQG 360 NGQRRKRKRA RSNPQQTQLS LNNQQPDDEA RNTLPDINNS NVLFAGHITN ESQPEKDQPE 420 LQLRDSNLSV VPSTNAVSTE DAFIGNGPSI NPVLENSEVV PYESRFHLGT QDSVVQHQLH 480 DTQLQSRTQF SGMNNEPRSS IFHYVPPNNG LHNGPENSVI HHELQDPGFA HGSQYPNLHQ 540 PHIYQYYTPS AEFVSAHDEQ QSHLAFNEPQ IKTKDSGVRS SVLHGSRNDI AREDHHYGKD 600 TFQHDHDRPV EMHFASPVAG GSPDYARLSS PFNFDLDVPS TLDTGDFELF LDEDVMTFFA 660 S |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 7e-73 | 167 | 298 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 363 | 368 | RRKRKR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
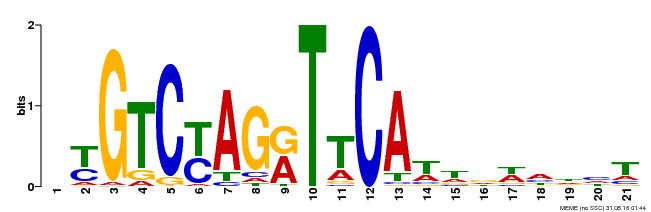
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975440 | 0.0 | HG975440.1 Solanum pennellii chromosome ch01, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009788577.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | ||||
| Refseq | XP_016465525.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-179 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A1S3ZLW2 | 0.0 | A0A1S3ZLW2_TOBAC; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A1U7X8Z3 | 0.0 | A0A1U7X8Z3_NICSY; ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | ||||
| STRING | XP_009788577.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA304 | 23 | 178 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-166 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



