 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009788810.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 826aa MW: 92878.5 Da PI: 6.9298 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 54.2 | 2.8e-17 | 2 | 46 | 74 | 118 |
CG-1 74 KvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
+vg+ e +++yYah+e++ptf rrcywlL+++le+ivlvhy+e++
XP_009788810.1 2 QVGNEERIHVYYAHGEDHPTFVRRCYWLLDKSLEHIVLVHYRETQ 46
699***************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01076 | 9.0E-4 | 1 | 46 | IPR005559 | CG-1 DNA-binding domain |
| PROSITE profile | PS51437 | 32.457 | 1 | 51 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.2E-14 | 2 | 44 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 8.8E-4 | 270 | 367 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 7.28E-12 | 278 | 364 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 1.48E-16 | 463 | 579 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.9E-16 | 463 | 577 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.4E-6 | 464 | 543 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.47E-12 | 479 | 573 | No hit | No description |
| PROSITE profile | PS50297 | 15.273 | 481 | 585 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 3.7E-6 | 514 | 543 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.728 | 514 | 546 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.95E-6 | 626 | 727 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.181 | 661 | 687 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 16 | 676 | 698 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.346 | 677 | 706 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.18 | 679 | 697 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 5.3E-4 | 699 | 721 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.889 | 700 | 724 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.0E-4 | 701 | 721 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 15 | 778 | 800 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.389 | 780 | 808 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 826 aa Download sequence Send to blast |
MQVGNEERIH VYYAHGEDHP TFVRRCYWLL DKSLEHIVLV HYRETQEAQG SPATSVAKGS 60 PATPVNSNSS SDPSDPSGWV LSEECNSVDE RTYGSSQHAH LEPNRDVTAK NHEQRLLEIN 120 TLEWDELLAP DNPNKLIATQ EAGGRASVGQ QNQIEVNGYS LNDGSLSVSR VPVASLESFV 180 CQVAGSDTVN FNPSNDMPFR SGDGQMTSNF RKNEPGVTTV GAGDSFDSLN KDGLQTQDSF 240 GRWINYFISD SPGSADEMMT PESSVTIDQS YVMQQIFNIT EISPTWALSS EETKILVIGH 300 FPGAQSQLAK SNLFCVCADV CFPAEFVQSG VYRCVISPQP PGLVSLYLSF DGNTPISQVM 360 TYEFRAPSAC KWTAPLEEQS SWDEFRVQMR LAHLLFSTSK SLSIFSSKVH QDSLKEAKRF 420 VRKCSHITDN WAYLIKSIED RKLPVPHAKD CLFELSLQTK FHEWLLERVI GGCKTSEWDE 480 QGQGVIHLCA ILGYTWAVYP FSWSGLSLDY RDKYGWTALH WAAHYGREKM VATLLSAGAK 540 PNLVTDPTSE NPGGSTAADL ASKNGFEGLG AYLAEKALVA HFKDMTLAGN VSGSLQTTTE 600 HINPGNFTEE ELYLKDTLAA YRTAADAAAR IQAAFREHSF KVQTKAVESS NPEMEARNIV 660 AAMKIQHAFR NYESRKKLAA AARIQYRFRS WKMRKDFLNM RRHAIKIQAV FRGFQVRKQY 720 RKIVWSVGVL EKAVLRWRLK RKGFRGLQVQ SSQAVDIKPD GDVEEDFFRA SRKQAEERVE 780 RSVVRVQAMF RSKRAQEEYR RMKLEHDNAT LEYERASLLN PDIQIG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
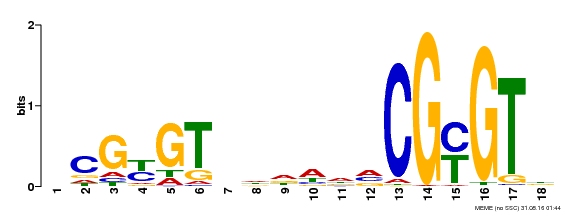
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN566049 | 0.0 | JN566049.1 Solanum lycopersicum calmodulin-binding transcription factor SR3 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009788810.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A1U7XQQ2 | 0.0 | A0A1U7XQQ2_NICSY; calmodulin-binding transcription activator 5 isoform X2 | ||||
| STRING | XP_009788809.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



