 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_009800960.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 442aa MW: 49305.4 Da PI: 8.101 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.7 | 2.7e-18 | 25 | 71 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT eEd++++++v+++G+ W+ Ia+ ++ gR +kqc++rw+++l
XP_009800960.1 25 KGPWTREEDDKIIELVAKYGPMKWSVIAKSLP-GRIGKQCRERWHNHL 71
79******************99**********.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 57.5 | 3e-18 | 77 | 120 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WT eE+ l+da++ +G++ W+ Ia+ ++ gRt++ +k++w++
XP_009800960.1 77 KDAWTLEEERALIDAHRIHGNK-WAEIAKVLP-GRTDNAIKNHWNS 120
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 4.286 | 1 | 19 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-6 | 4 | 27 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 31.737 | 20 | 75 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.65E-31 | 22 | 118 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.8E-17 | 24 | 73 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-17 | 25 | 71 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.73E-15 | 27 | 71 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-27 | 28 | 78 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 23.595 | 76 | 126 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.7E-18 | 76 | 124 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.6E-16 | 77 | 120 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.31E-13 | 79 | 122 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-23 | 79 | 126 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 442 aa Download sequence Send to blast |
MLEKNRSEVQ CLHRWQKVLN PDLVKGPWTR EEDDKIIELV AKYGPMKWSV IAKSLPGRIG 60 KQCRERWHNH LNPNIKKDAW TLEEERALID AHRIHGNKWA EIAKVLPGRT DNAIKNHWNS 120 SLRKKLDFYL ATGNLPPATR DGLQNGCKDT TETAKAEESL VASSGTADMC KIVNGWQVTE 180 ISASNGGPQT ESTDSEVVRL EAQSPEIDAI RHIKPITESE TLYEGRRINS VVDRLKVVEA 240 PFHCEIPTCG TLYYEPPTSE SCISLDSDLL NIRWAQCESD ASPSPSPNGF FTPPSTKGSS 300 LYAQTPESVL KIAARSFPNT PSILRKRKTQ AKLSTPTNKM GKSDGDFCKD KPTDACEEIL 360 PRNSSNGIGL YNSQAFNASP PYRLRSKRTS IFKSVEKQLE FALNNEKHDT SNGISESTVN 420 DISHAAKDFS QSAKKDTDKK VE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 5e-63 | 5 | 126 | 38 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 5e-63 | 5 | 126 | 38 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
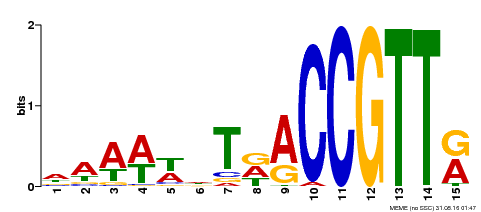
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975448 | 0.0 | HG975448.1 Solanum pennellii chromosome ch09, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009800960.1 | 0.0 | PREDICTED: myb-related protein B-like isoform X3 | ||||
| Swissprot | Q6R032 | 1e-107 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | A0A1U7YQ36 | 0.0 | A0A1U7YQ36_NICSY; myb-related protein B-like isoform X3 | ||||
| STRING | XP_009800958.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-109 | myb domain protein 3r-5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



