 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010086977.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 399aa MW: 43877.6 Da PI: 7.3495 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 134.2 | 4.1e-42 | 95 | 171 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
C v+gC+adls++++yhrrhkvCe hsk+p+v++ g++qrfCqqCsrfh+l+efDe+krsCr+rL++hn+rrrk+q+
XP_010086977.1 95 CLVDGCNADLSNCRDYHRRHKVCELHSKTPQVTIGGHKQRFCQQCSRFHSLEEFDEGKRSCRKRLDGHNRRRRKPQP 171
**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.3E-34 | 88 | 156 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.231 | 92 | 169 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.49E-39 | 93 | 174 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.6E-31 | 95 | 168 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 399 aa Download sequence Send to blast |
MDWNSKAPTS WDFTELEQET FSNLDTVNGS NSFGDQKAIR EDFSVDLKLG QTGNSGNEMV 60 DTWREPGAVP KPVSSPSGSS KRSRGSYNGS QAVSCLVDGC NADLSNCRDY HRRHKVCELH 120 SKTPQVTIGG HKQRFCQQCS RFHSLEEFDE GKRSCRKRLD GHNRRRRKPQ PEPFSRYGNL 180 LSNYTGTQLL PFSSSLIYPS TTVVNPSWPG GVVVKTEADS GLHNHHPQLP FLDKQNMFMG 240 SPSPTTNTTY KGCSNKPFPL LQDHSPTINS SHTAPEVSAC HNHPLLRTFP LCESSSESKS 300 KAFFDVQHSD CALYLLSSSQ AQTSEISLGH NNSQLGSMPM VHHPALYVHG GLEPMSSVLM 360 ANGSSGNAES VHCPNGMFHM GSVGNEAPHP HETLPSHWG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-28 | 85 | 168 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 151 | 168 | KRSCRKRLDGHNRRRRKP |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00555 | DAP | Transfer from AT5G50570 | Download |
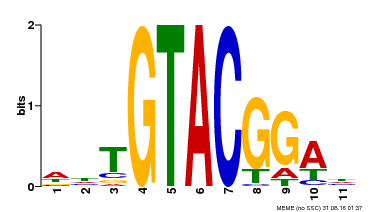
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010086977.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010086977.1 | 0.0 | squamosa promoter-binding-like protein 13A | ||||
| Refseq | XP_024017871.1 | 0.0 | squamosa promoter-binding-like protein 13A | ||||
| TrEMBL | W9QLM6 | 0.0 | W9QLM6_9ROSA; Squamosa promoter-binding-like protein 13 | ||||
| STRING | XP_010086977.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2591 | 33 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50670.1 | 2e-69 | SBP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21393429 |



