 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010088954.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | NF-YB | ||||||||
| Protein Properties | Length: 175aa MW: 19726.8 Da PI: 5.0866 | ||||||||
| Description | NF-YB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NF-YB | 163 | 4e-51 | 5 | 100 | 2 | 97 |
NF-YB 2 reqdrflPianvsrimkkvlPanakiskdaketvqecvsefisfvtseasdkcqrekrktingddllwalatlGfedyveplkvylkkyrelege 96
+eqdr+lPianv+rimk++lP akisk+ak+t+qec++efisfvt+easdkc++e+rkt+ngdd++wal+ lGf++y+e l yl+kyre+e+e
XP_010088954.1 5 QEQDRMLPIANVGRIMKQILPPTAKISKEAKQTMQECATEFISFVTGEASDKCHKENRKTVNGDDICWALTALGFDNYAEVLVRYLHKYREAERE 99
79*******************************************************************************************99 PP
NF-YB 97 k 97
k
XP_010088954.1 100 K 100
7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.20.10 | 4.9E-51 | 4 | 132 | IPR009072 | Histone-fold |
| SuperFamily | SSF47113 | 5.78E-40 | 7 | 129 | IPR009072 | Histone-fold |
| Pfam | PF00808 | 1.4E-26 | 10 | 74 | IPR003958 | Transcription factor CBF/NF-Y/archaeal histone domain |
| PRINTS | PR00615 | 5.4E-18 | 38 | 56 | No hit | No description |
| PROSITE pattern | PS00685 | 0 | 41 | 57 | IPR003956 | Transcription factor, NFYB/HAP3, conserved site |
| PRINTS | PR00615 | 5.4E-18 | 57 | 75 | No hit | No description |
| PRINTS | PR00615 | 5.4E-18 | 76 | 94 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 175 aa Download sequence Send to blast |
MVDQQEQDRM LPIANVGRIM KQILPPTAKI SKEAKQTMQE CATEFISFVT GEASDKCHKE 60 NRKTVNGDDI CWALTALGFD NYAEVLVRYL HKYREAEREK ANQISKGSDN NNNNNHITIA 120 DHDEDKDGDS DDRSKSTTGV QQTDQISNII TTSPTLMEFN IIEKGDHHQS LVKSS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4g91_B | 2e-42 | 5 | 95 | 2 | 92 | Transcription factor HapC (Eurofung) |
| 4g92_B | 2e-42 | 5 | 95 | 2 | 92 | Transcription factor HapC (Eurofung) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the NF-Y/HAP transcription factor complex. The NF-Y complex stimulates the transcription of various genes by recognizing and binding to a CCAAT motif in promoters. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00133 | DAP | Transfer from AT1G09030 | Download |
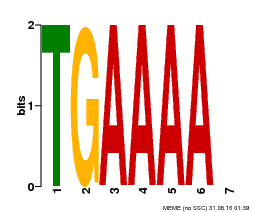
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010088954.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010088954.1 | 1e-130 | nuclear transcription factor Y subunit B-4 | ||||
| Swissprot | O04027 | 2e-54 | NFYB4_ARATH; Nuclear transcription factor Y subunit B-4 | ||||
| TrEMBL | W9QH19 | 1e-128 | W9QH19_9ROSA; Nuclear transcription factor Y subunit B-4 | ||||
| STRING | XP_010088954.1 | 1e-129 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF805 | 32 | 131 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09030.1 | 2e-56 | nuclear factor Y, subunit B4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21396705 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



