 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010089258.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 364aa MW: 40139.8 Da PI: 8.5221 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 132 | 2e-41 | 211 | 288 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cq+egC+adls+ak+yhrrhkvCe+hska++v+++gl+qrfCqqCsrfh lsefD++krsCr+rLa+hn+rrrk+q+
XP_010089258.1 211 RCQAEGCNADLSHAKHYHRRHKVCEFHSKASTVVAAGLTQRFCQQCSRFHLLSEFDNGKRSCRKRLADHNRRRRKTQQ 288
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.2E-35 | 204 | 273 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.419 | 209 | 286 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.27E-38 | 210 | 289 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 7.0E-31 | 212 | 285 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009554 | Biological Process | megasporogenesis | ||||
| GO:0009556 | Biological Process | microsporogenesis | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 364 aa Download sequence Send to blast |
MLDYEWGNPS AIMLAGEEPE QHPGSDPTRQ NIIEHYASAQ AAFNDHNAHF LTPHLQSHNH 60 NHHHHHHHHE HANNNDNNPS AFPHHHFGIA TAHHHTQAQP QQQQLHVHHH SSLYDVPRAY 120 ASASSYAPAS ILSLDPITAG GGGGGGGGLG VPFMLVPKSE DHQRAAVDFS SRIGLNLGGR 180 TYFSSEDDFM SRLYRRPRAL EAGSTSANSP RCQAEGCNAD LSHAKHYHRR HKVCEFHSKA 240 STVVAAGLTQ RFCQQCSRFH LLSEFDNGKR SCRKRLADHN RRRRKTQQPN QQVQNPKSLQ 300 PENAQNPSSD NVARSPPDSG AQSSSSVTVA VSPPRMSLDC FRQRSYQTTT ASSGSSSSLF 360 FSSG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-30 | 203 | 285 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
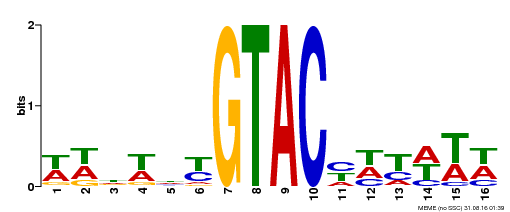
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. Binds specifically to the 5'-GTAC-3' core sequence. Involved in development and floral organogenesis. Required for ovule differentiation, pollen production, filament elongation, seed formation and siliques elongation. Also seems to play a role in the formation of trichomes on sepals. May positively modulate gibberellin (GA) signaling in flower. {ECO:0000269|PubMed:12671094, ECO:0000269|PubMed:16095614, ECO:0000269|PubMed:17093870}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00090 | SELEX | Transfer from AT1G02065 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010089258.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010089258.1 | 0.0 | squamosa promoter-binding-like protein 8 | ||||
| Swissprot | Q8GXL3 | 1e-85 | SPL8_ARATH; Squamosa promoter-binding-like protein 8 | ||||
| TrEMBL | W9QQ44 | 0.0 | W9QQ44_9ROSA; Squamosa promoter-binding-like protein 8 | ||||
| STRING | XP_010089258.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6801 | 33 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.1 | 9e-72 | squamosa promoter binding protein-like 8 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21404629 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



