 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010089911.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 496aa MW: 53978 Da PI: 7.1497 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 126.5 | 7.9e-40 | 127 | 187 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++k+l+cprC+s++tkfCyynny+++qPr+fCkaC+ryWt+GG++rnvPvG+grrknk+ss
XP_010089911.1 127 PDKILPCPRCNSMDTKFCYYNNYNVNQPRHFCKACQRYWTAGGTMRNVPVGAGRRKNKNSS 187
7899******************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 1.0E-31 | 120 | 184 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 2.7E-32 | 129 | 185 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.863 | 131 | 185 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 133 | 169 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 496 aa Download sequence Send to blast |
MLESKDPAIK LFGKKIAVPA DDEARAVSGE ELASQPLEKE DVNEEETETE MKDSPPRSAS 60 ENSGQDSDPP SETEDSINPE TLPDSSNVNP KTPSVNEESA KSNIAKIDNE KNNASAANAQ 120 EKTLKKPDKI LPCPRCNSMD TKFCYYNNYN VNQPRHFCKA CQRYWTAGGT MRNVPVGAGR 180 RKNKNSSSHY RHITISEALQ AARIEAAPPN GTHPHHHHHH HHHHHPALKS NGRVLSFALD 240 APICDSMASV LNLTDNKKAP RNGFHKTEER GFPGPCKGRE NGDDCSSASS ITVSNSMDEV 300 GRNSTQEPQV HNMNGFSSIP CIPGVPWPYP WNSAVPPPPP PPPPFCPPGF PMSFYPAAYW 360 NCAVPGTWNI PWFAPPQPSS PSQNSPSLGS NSPTLGKHSR DGEILKSDSL EKEEPSKPKN 420 GSVLVPKTLR IDDPTEAAKS SIWATLGIKN DGISGGGMFK AFQSKSDEKK SMAETSPVLR 480 ANPAALSRSL KFHESS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Regulates a photoperiodic flowering response. Transcriptional repressor of 'CONSTANS' expression. {ECO:0000250, ECO:0000269|PubMed:19619493}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00394 | DAP | Transfer from AT3G47500 | Download |
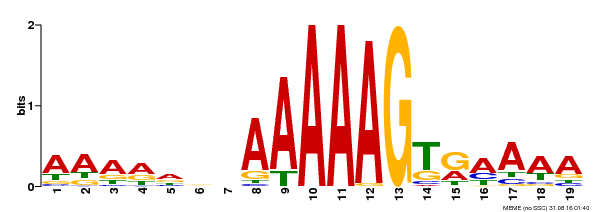
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010089911.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Highly expressed at the beginning of the light period, then decreases, reaching a minimum between 16 and 29 hours after dawn before rising again at the end of the day. {ECO:0000269|PubMed:19619493}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010089911.1 | 0.0 | cyclic dof factor 3 | ||||
| Swissprot | Q8LFV3 | 1e-117 | CDF3_ARATH; Cyclic dof factor 3 | ||||
| TrEMBL | W9QRU1 | 0.0 | W9QRU1_9ROSA; Dof zinc finger protein | ||||
| STRING | XP_010089911.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF625 | 34 | 147 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47500.1 | 1e-102 | cycling DOF factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21391960 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



