 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010090255.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 700aa MW: 79571.8 Da PI: 6.4167 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 49.9 | 8.5e-16 | 123 | 185 | 2 | 69 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegek 69
W+++evlaL+++r+ me+++ + We+vs+k++e gf+rs ++Ckek+e+ ++++ +i++ k
XP_010090255.1 123 WSNDEVLALLRIRSTMENWFPDF-----TWEHVSRKLSEIGFKRSGEKCKEKFEEESRYFNNINNCTK 185
********************998.....9********************************9998665 PP
| |||||||
| 2 | trihelix | 92.5 | 4.3e-29 | 528 | 620 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgk........lkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+++evlaLi++r ++ ++ + k+plWe++s+ m+e g++rs+k+Ckekwen+nk+++k+k+ +kkr s +s+tcpyf+ql
XP_010090255.1 528 RWPRDEVLALINLRCSLYNSGDHIQekegsnsvVKAPLWERISQGMSELGYKRSAKRCKEKWENINKYFRKTKDVNKKR-SVESRTCPYFHQLS 620
8**************6666665322223445559********************************************8.67778*******95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 6.342 | 115 | 174 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 13 | 119 | 176 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 7.6E-10 | 121 | 183 | No hit | No description |
| SMART | SM00717 | 0.5 | 525 | 595 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 1.6E-18 | 527 | 620 | No hit | No description |
| CDD | cd12203 | 3.57E-24 | 527 | 600 | No hit | No description |
| PROSITE profile | PS50090 | 6.353 | 528 | 593 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 700 aa Download sequence Send to blast |
MFDGVPPHDQ LHQFIASSRA AAAASPLPLP LSSSPLHSQT AFSTNSFDVS AYNIIPHHHQ 60 LLLPHQQHQQ PQFVHPILLH QSSNNNSHKS SINNEEKEAS NLVEIERERS VPEPNNTIDD 120 PAWSNDEVLA LLRIRSTMEN WFPDFTWEHV SRKLSEIGFK RSGEKCKEKF EEESRYFNNI 180 NNCTKSCRFL CTDHLEVEEE EEVEEGLNYH TQNPAHQTNI IGTQKRQIIS VVLPENQDKV 240 RGATGTGNGT AQISLEDQLG SPRNHDHDHH QTFNGSAKES DHDLENDHHD SSSTRSKSKK 300 RKRQKIKFEM FKGFCEDIVS KMMSQQEEMH NKLVEDMVKR DKEKVEKEEA WKKQEMDRMN 360 KELDIMAHEQ AIAGDRQATI IDFLNKFISS SSSGTITTTT TTISSESDHH INHFSGPKKG 420 KDSLDKAVLV KNTPKNHTDF SLTPLIQAAH GSLNDLEEQN NSNIVVEEQP SSSAMIVRPQ 480 NSSLSTQKNV ISEQPQQNPT SSPTKNPSST TSTQKVAIND IKDDVGKRWP RDEVLALINL 540 RCSLYNSGDH IQEKEGSNSV VKAPLWERIS QGMSELGYKR SAKRCKEKWE NINKYFRKTK 600 DVNKKRSVES RTCPYFHQLS TLYNKGTLVA PNSQGPDQEM NHHSSQLESL ADHDHDDHDD 660 DDHHHHSSDV HVAEGERSPH NHVVVVHHQE VVPAAFDFEF |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
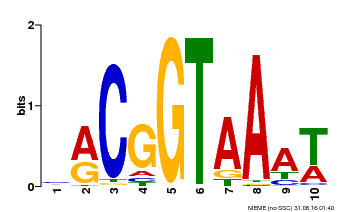
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010090255.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015041 | 1e-37 | AP015041.1 Vigna angularis var. angularis DNA, chromosome 8, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010090255.1 | 0.0 | trihelix transcription factor GTL2 isoform X2 | ||||
| TrEMBL | W9R6Q3 | 0.0 | W9R6Q3_9ROSA; Trihelix transcription factor GTL2 | ||||
| STRING | XP_010090255.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5043 | 33 | 55 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 1e-56 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21389601 |



