 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010090934.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 322aa MW: 36140.3 Da PI: 6.0302 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 60.8 | 3.2e-19 | 30 | 79 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++ GVr+++ +gr++AeIrdps+ + r++lg+f+taeeAa a+++a+++++g
XP_010090934.1 30 RFLGVRRRP-WGRYAAEIRDPST---KERHWLGTFDTAEEAALAYDRAARSMRG 79
688******.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 7.05E-31 | 29 | 86 | No hit | No description |
| Pfam | PF00847 | 3.5E-12 | 30 | 79 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.9E-29 | 30 | 87 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.3E-34 | 30 | 93 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.719 | 30 | 87 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 5.43E-21 | 30 | 88 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 4.4E-8 | 31 | 42 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.4E-8 | 53 | 69 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0010030 | Biological Process | positive regulation of seed germination | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 322 aa Download sequence Send to blast |
MSSNINNSNN KMKKKQDLVQ AGTDDKTSPR FLGVRRRPWG RYAAEIRDPS TKERHWLGTF 60 DTAEEAALAY DRAARSMRGS RARTNFVYSD MPPGSSVTSI LSPDDQNGHL FVNVAAAAGH 120 DHDDRRRHQQ LGGLVTQAPP PDPTPQPQLL YDHQDEAMMM MMMSLGEANG IHRAQAQVIM 180 SNNNYQHENY NYISNDHDED HHRHPMAIIN SSSSELPPFP ASEFNSDDQY HYYNCNTAST 240 SNSNSSNIMQ AGAGAMNMMM QDSSCEDQYG YNYSYNSNYV HSPLFSTMPS VSNSDHHHSQ 300 SQSQFPDPFD LGSTTTSSSF FF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 1e-20 | 31 | 86 | 3 | 59 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00509 | DAP | Transfer from AT5G13910 | Download |
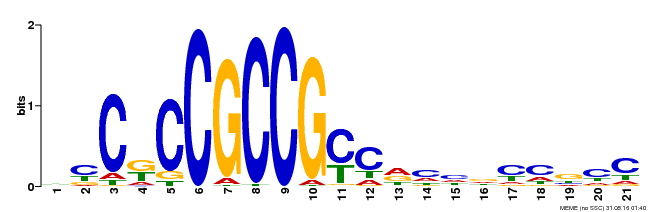
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010090934.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010090934.2 | 0.0 | ethylene-responsive transcription factor LEP | ||||
| TrEMBL | W9QM18 | 0.0 | W9QM18_9ROSA; Ethylene-responsive transcription factor LEP | ||||
| STRING | XP_010090934.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3904 | 31 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13910.1 | 2e-29 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21390964 |



