 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010093193.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 512aa MW: 56763.5 Da PI: 7.0244 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52.7 | 7.7e-17 | 318 | 364 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+ksLq
XP_010093193.1 318 VHNLSERRRRDRINEKMKALQELIPHS-----NKTDKASMLDEAIEYLKSLQ 364
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 3.01E-20 | 312 | 378 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.254 | 314 | 363 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.53E-18 | 317 | 368 | No hit | No description |
| Pfam | PF00010 | 3.1E-14 | 318 | 364 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 8.7E-20 | 318 | 372 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.9E-18 | 320 | 369 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 512 aa Download sequence Send to blast |
MNNCISDWNF EGDLPINGQK NPTGGDHDLV EILWRNGQVV LHSQTHRKQA LNNPNDPRQV 60 QKHDHHDQQA TRACGSYGNS INMSQDDESA SWIHYPLEDS FEKEFCSNLF SDSPSCDPID 120 IDKVPIIRQF EEEKVSKFGA YETTTHVKGL KPSCHENQML PPRQYCNSAL QDHNMGSLGK 180 VVNFSQFSGP EKCDLRPSGG QGEGRECSVM TVGLSHCGSN QLPNDHDMIR KVIPQSESGK 240 TVTLEPTVTS SSGGSGSSLG RPCKQSTTTT SGNTNNKRKS RDEEELECQS KAAELESAVA 300 NKSSQRSGTL RKSRAAEVHN LSERRRRDRI NEKMKALQEL IPHSNKTDKA SMLDEAIEYL 360 KSLQLQLQVM WMGSGMAPMM FPGVQHYMSR MGMAMAPPAL PSIHNPMHLP RVPVVDQSML 420 VAPTTDQSVL CQTPAFNPLN YQNQMQNTSF PEQFARYMGF HSMQPVSQPL NIFRFSPQTV 480 QQSQSMVQHG NSTGPSNSGF PPDGGISGKM GK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 322 | 327 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
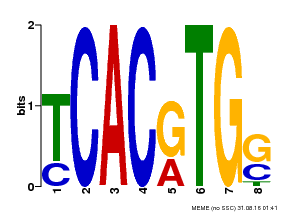
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010093193.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010093193.2 | 0.0 | transcription factor PIF4 isoform X1 | ||||
| TrEMBL | W9QT82 | 0.0 | W9QT82_9ROSA; Uncharacterized protein | ||||
| STRING | XP_010093193.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8146 | 29 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43010.2 | 4e-66 | phytochrome interacting factor 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21396494 |



