 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010093403.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 378aa MW: 41332.4 Da PI: 8.2972 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.3 | 9.2e-14 | 75 | 123 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f ++eAa+a++ a+++++g
XP_010093403.1 75 SKYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEDEAARAYDVAAQRFRG 123
89****9888.8*********3.....5**********99*************98 PP
| |||||||
| 2 | B3 | 99.3 | 2.3e-31 | 207 | 306 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefe 91
f+k+ tpsdv+k++rlv+pk++ae+h + ++ +++ l +ed g++W+++++y+++s++yvltkGW++Fvk++ Lk+gD+v F+++ +++
XP_010093403.1 207 FEKAVTPSDVGKLNRLVIPKQHAEKHfplqSANTCKGVLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKSLKAGDIVSFQRSTGPDKQ 301
89*************************9986677799**************************************************87667777 PP
.EEEE CS
B3 92 lvvkv 96
l++ +
XP_010093403.1 302 LYIDW 306
77766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.57E-17 | 75 | 131 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.9E-9 | 75 | 123 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.29E-26 | 75 | 130 | No hit | No description |
| PROSITE profile | PS51032 | 20.455 | 76 | 131 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 7.7E-21 | 76 | 131 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.2E-30 | 76 | 137 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 3.7E-41 | 202 | 310 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.35E-29 | 206 | 302 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 3.91E-29 | 206 | 295 | No hit | No description |
| Pfam | PF02362 | 1.3E-28 | 207 | 306 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.4E-24 | 207 | 310 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.029 | 207 | 310 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MDGNSSTDES TTTESFTLSA SPAATPTPFK SPPAAESTLC RVGSGASSVV VDSEMITNAI 60 NGGADAESGR KLPSSKYKGV VPQPNGRWGA QIYEKHQRVW LGTFNEEDEA ARAYDVAAQR 120 FRGRDAVTNF KPLSETTEED EIESVFLNSH SKAEIVDMLR KHTYNDELEQ SKRNSFGIDG 180 NGNRKPSRTG LLFGSDLTAS MAREQLFEKA VTPSDVGKLN RLVIPKQHAE KHFPLQSANT 240 CKGVLLNFED VGGKVWRFRY SYWNSSQSYV LTKGWSRFVK EKSLKAGDIV SFQRSTGPDK 300 QLYIDWKARD VDSGSAGGAV GPVQMVRLFG VNIFKLPGSE MVEGIGIGIG GGCNGKRTRD 360 MELLELECSK KPRIIGAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 2e-60 | 204 | 322 | 11 | 127 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds specifically to bipartite recognition sequences composed of two unrelated motifs, 5'-CAACA-3' and 5'-CACCTG-3'. May function as negative regulator of plant growth and development. {ECO:0000269|PubMed:15040885, ECO:0000269|PubMed:9862967}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
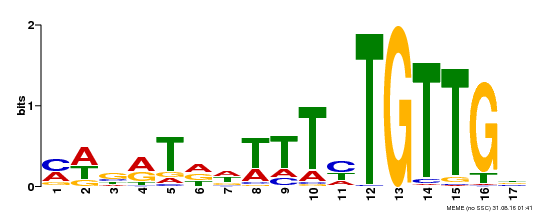
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010093403.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by brassinosteroid and zeatin. {ECO:0000269|PubMed:15040885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU732460 | 4e-58 | GU732460.1 Malus x domestica AP2 domain class transcription factor (AP2D36) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010093403.1 | 0.0 | AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| Swissprot | Q9ZWM9 | 1e-146 | RAV1_ARATH; AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| TrEMBL | W9QTI0 | 0.0 | W9QTI0_9ROSA; AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| STRING | XP_010093403.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2920 | 32 | 74 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13260.1 | 1e-138 | related to ABI3/VP1 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21405755 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



