 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010094157.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1064aa MW: 119574 Da PI: 6.5159 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 126 | 1.5e-39 | 56 | 130 | 44 | 118 |
CG-1 44 rkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
++++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+een++fqrrcyw+Lee+l +iv+vhylevk
XP_010094157.1 56 HQVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEENENFQRRCYWMLEEDLMHIVFVHYLEVK 130
4789********************************************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 54.566 | 25 | 135 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.1E-42 | 33 | 130 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 9.1E-33 | 57 | 129 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.1E-5 | 451 | 552 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 5.9E-6 | 467 | 546 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 2.0E-14 | 467 | 552 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 2.8E-15 | 650 | 774 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.769 | 651 | 783 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.2E-15 | 652 | 773 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.34E-12 | 657 | 771 | No hit | No description |
| PROSITE profile | PS50088 | 8.763 | 668 | 700 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.19E-8 | 879 | 938 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.15 | 887 | 909 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.187 | 888 | 917 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0017 | 889 | 908 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 910 | 932 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.285 | 911 | 935 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.7E-4 | 913 | 932 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1064 aa Download sequence Send to blast |
MGNKMRKKEE GKRGTKMICN NYYQRRNIVG YAQLKFVKFF VTIKSFIFHQ SPQTGHQVLR 60 YFRKDGHNWR KKKDGKTVKE AHEKLKVGSI DVLHCYYAHG EENENFQRRC YWMLEEDLMH 120 IVFVHYLEVK GNRTNIGGIR EIDEVSQSLR EGSPQTSSSS TSHYKAPSGS TDYTSPSSTL 180 TSLCEDADSG DIHRASSRLH SYLESPQLQL KMDMGSVHPN SNLREGHLGQ SSIHGANYVP 240 HFQGDRPNYS EPATCATGYQ NTLGLGSWEE ILEQCTTGFN TVPSHVSVST SQPACTGVVH 300 EQENLVSGRL LAGESITKEE LGNSLSNYST WQISLEDNLL PLPKGLVEQS SNLEMPYDLE 360 NMLFENSTAD SSLTSSPYQL DSHLDQQTEN STEGNINYAF TLRQQLLDGE EGLKKLDSFS 420 RWVTKELGEV DDLQMQSSSG IPWSTVENVV DDSSLSPSIS QDQLFSIIDF SPKWGFTDSQ 480 PKVLIIGTFL KSRQEVEKYK WSCMFGEEEG PAEGLADGIL CCYAPPHTAG PVPFYVTCSN 540 RLACSEVREF DYKCGATKDL DIRDIYNDNT VELRLHMRLE GLLHLGSVNP TSFSFRSTVE 600 KRTLISKIIS LKEEEESHQK VDQADEKDLS QYKVKEHLFT KLMKEKLYSW LLQKATEDGK 660 GPNILDDEGQ GVLHLAAALG YNWAIKPIVT AGVSINFRDI NGWTALHWAA FYGRQGLLNL 720 KLSFAEHTVA FLVSQGAASG AVTDPSPEFP LGRSAADLAS VNGHKGISGF LAESSLTSHL 780 SSLSVNDSKE DGGAEISGTK AVQTVSERKA TPVTYGEMPD ALSLKDSLTA VRNATQAADR 840 IHQMFRMQSF ERKQLNEYDD DGCGLSDERA LSLLAGRSRK SGQNDRLAHS AAVQIQKKFR 900 GWKKRKEFLL IRQRIVKIQA HVRGHQVRKQ YRAIVWSVGI LDKVILRWRR KGSGLRGFRP 960 DAIPKEPKLP SMPIKEDEYD FFKEGRKQTE ERLQKALTRV KSMVQYPEGR AQYRRVLNVV 1020 QGLQETKVTD MALIDSEEIA DADDDVIKID QFLDDDTFMS IAFE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
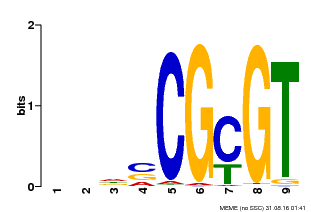
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010094157.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024019885.1 | 0.0 | calmodulin-binding transcription activator 2 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | W9RHV9 | 0.0 | W9RHV9_9ROSA; Calmodulin-binding transcription activator 2 | ||||
| STRING | XP_010094157.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7406 | 26 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21400207 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



