 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010094499.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 331aa MW: 36525.1 Da PI: 4.5513 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 32.1 | 2.4e-10 | 160 | 201 | 4 | 45 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
+k+++r +NR AA rsR+RKk +++ Le k+k Le+e +++
XP_010094499.1 160 SKKQKRQLRNRDAAVRSRERKKLYMKDLEMKSKYLEGECRRM 201
79***********************************98766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 3.4E-10 | 153 | 219 | No hit | No description |
| SMART | SM00338 | 2.0E-4 | 154 | 221 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.255 | 159 | 201 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 3.8E-9 | 160 | 201 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 5.06E-10 | 161 | 217 | No hit | No description |
| CDD | cd14704 | 1.21E-15 | 162 | 213 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 164 | 179 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MEGGDVEFWG GEDVIREIDW SSIFNDGFHG GANILEWDPP AVITDSSASV SSSSSSPKAD 60 AFLKNSPDSV PSWTDGIEKL LMEDDDDNKV SPEPSSDYCE RFLVDLLVDS PADVSGEAVA 120 ASTNSDPNVT ADAYANSEEE EEKEKVEAAE VDDEADEPIS KKQKRQLRNR DAAVRSRERK 180 KLYMKDLEMK SKYLEGECRR MGRLLQCCYA ENQALRVTLQ MGGAFGASAT KQESAVLLLE 240 SLLLGSLLWL VVITCLFIPP AILEAVLMRE DKAKRAPERV APREAGGTRS EIFGYAVGQR 300 QSGFVKSRRR TCKASWTKMK HFPCLSPLAL V |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
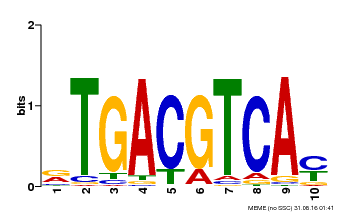
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010094499.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010094499.1 | 0.0 | bZIP transcription factor 60 | ||||
| TrEMBL | W9QWD5 | 0.0 | W9QWD5_9ROSA; BZIP transcription factor 60 | ||||
| STRING | XP_010094499.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF12009 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 6e-19 | basic region/leucine zipper motif 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21407516 |



