 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010095053.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 294aa MW: 33640.7 Da PI: 7.365 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 29.3 | 2.1e-09 | 26 | 70 | 3 | 45 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwq 45
WT+eE +++ a + + +W+++a++++ g++ ++++ +
XP_010095053.1 26 EWTKEENKKFESALAIYDETtpnRWAKVAEMIP-GKSVYDVIKQYK 70
7*****************99*************.*******98886 PP
| |||||||
| 2 | Myb_DNA-binding | 38.1 | 3.6e-12 | 128 | 172 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ +++ + ++G+g+W+ I+r + ++t+ q+ s+ qky
XP_010095053.1 128 PWTKDEHRQFLLGLLKYGKGDWRNISRNFVVTKTPTQVASHAQKY 172
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 10.253 | 22 | 76 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.3E-10 | 23 | 75 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 8.22E-12 | 25 | 79 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-4 | 26 | 69 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.12E-8 | 26 | 72 | No hit | No description |
| Pfam | PF00249 | 7.3E-8 | 26 | 71 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.332 | 121 | 177 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.12E-15 | 123 | 177 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.1E-17 | 124 | 176 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.9E-10 | 125 | 175 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.2E-10 | 125 | 171 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.7E-10 | 128 | 172 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.12E-8 | 128 | 173 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 294 aa Download sequence Send to blast |
METLHPTWYL PNSSWLFQGI SQSSTEWTKE ENKKFESALA IYDETTPNRW AKVAEMIPGK 60 SVYDVIKQYK ELEEDVCDIE AGRVPIPGYF SPSDFTTIDL VDDRDDSFRK KPQSARASSD 120 HERKKGVPWT KDEHRQFLLG LLKYGKGDWR NISRNFVVTK TPTQVASHAQ KYFLRQHSGG 180 KDKRRPSIHD FTTVNLRESN TSPSENIINL PNFFDHSSVV HSEEAHKGLL DWNIPSEEAL 240 MVLGSTCGNL FMNSPYDISS PKLPLRQNLY GGGARFAAHT KPQSSMFEMQ TSRY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 5e-15 | 23 | 89 | 7 | 73 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00347 | DAP | Transfer from AT3G11280 | Download |
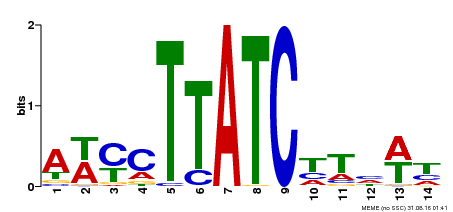
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010095053.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010095053.1 | 0.0 | transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 1e-87 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | W9R8G5 | 0.0 | W9R8G5_9ROSA; Transcription factor | ||||
| STRING | XP_010095053.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF466 | 34 | 162 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G11280.2 | 4e-90 | MYB family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21409021 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



