 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010095167.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 940aa MW: 103367 Da PI: 4.863 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 77.7 | 1.2e-24 | 143 | 244 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++p++ ae+ + ++++ ++l+++d++ +W++++iyr++++r++lt+GW+ Fv a++L++gD+v+F +++
XP_010095167.1 143 FCKTLTASDTSTHGGFSVPRRAAEKLfppldytM-QPPN-QELVVRDLHDTTWTFRHIYRGQPKRHLLTTGWSLFVGAKRLRAGDSVLFI--RDE 233
99*********************999*****954.5444.49************************************************..457 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
+ +l+v+v+r+
XP_010095167.1 234 KSQLMVGVRRA 244
7778*****97 PP
| |||||||
| 2 | Auxin_resp | 117.3 | 1.3e-38 | 269 | 352 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+++s+F+++YnPra++seFv++++k++ka++ +++svGmRf m+fete+s +rr++Gt+vg+sdldp+rWp+SkWr+L+
XP_010095167.1 269 AAHAAANRSPFTIFYNPRACPSEFVIPLAKYRKAVYaTQLSVGMRFGMMFETEESGKRRYMGTIVGISDLDPLRWPGSKWRNLQ 352
79*********************************989********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.31E-47 | 130 | 272 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 3.1E-42 | 136 | 257 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 6.48E-23 | 142 | 243 | No hit | No description |
| Pfam | PF02362 | 4.9E-22 | 143 | 244 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.14 | 143 | 245 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.4E-25 | 143 | 245 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 7.5E-34 | 269 | 352 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 25.929 | 826 | 910 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 1.77E-9 | 829 | 905 | No hit | No description |
| Pfam | PF02309 | 6.4E-7 | 871 | 912 | IPR033389 | AUX/IAA domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 940 aa Download sequence Send to blast |
MGTMEEKIKT GSPANLLEEM KLLKEMQQDQ SGSKNSINSE LWHACAGPLV SLPQVGSLVY 60 YFPQGHSEQV AVSTKRKATS QIPNYPNLPS QLMCQVQNIT LHADRDTDEI YAQMSLQPVN 120 SEKDVFPVPD FGLKPSKHPT EFFCKTLTAS DTSTHGGFSV PRRAAEKLFP PLDYTMQPPN 180 QELVVRDLHD TTWTFRHIYR GQPKRHLLTT GWSLFVGAKR LRAGDSVLFI RDEKSQLMVG 240 VRRANRQQST LPSSVLSADS MHIGVLAAAA HAAANRSPFT IFYNPRACPS EFVIPLAKYR 300 KAVYATQLSV GMRFGMMFET EESGKRRYMG TIVGISDLDP LRWPGSKWRN LQVEWDEPGC 360 CDKQNRVSPW EIETPESLFI FPSLTAGLKR PFHAGYLETE WGNMVKRPFI RVPENGSADL 420 PYSISNLYSE QLMKVLLKPQ LINYSGTLAS LQQEAAAKAD PPQDMKMQAT MNQKHPIVCS 480 ESLALQNQIS PQSSLDQSCV SNLNSSANAN NPPGNFNSAA KVEGRKVGGI STEKSKFENE 540 VSTDQLSQLA STEQGNDEKL AAGIANPQAI INQLTHLNQN QNPVQLQTSQ WGIQPPLESL 600 MYLSQQTEAM PSDITSTNVS LPSLDTDDCM FYPSCQPYAG LLRSPGPLSV FGLQDSSVFP 660 ESNNFPLPSI GQGMWDNHNL KVQPDQVPPF SQQDASNINC ISNSSSLRDL SDESNTQSGI 720 YSCQNIDGSN GGSIVVDPSV SSTILDDFST LKNVDFQNPS DCLVGNFSSS QDVQSQITSA 780 SLGDSQAFSR QDRQDNSGGT SSSNVDLDDS SLLQNSSWQQ VVPPVRTYTK VQKAGSVGRS 840 IDVSSFTNYD ELCAAIECMF GLEGLLNDPR GSGWKLVYVD YENDVLLVGD DPWEEFVGCV 900 RCIRILSPTE VQQMSEEGMK LLNSAAMQGI NGSILEAGRA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 1 | 377 | 5 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
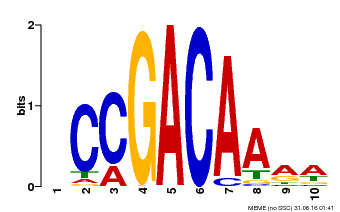
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010095167.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010095167.1 | 0.0 | auxin response factor 5 | ||||
| Refseq | XP_024020560.1 | 0.0 | auxin response factor 5 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | W9R8R1 | 0.0 | W9R8R1_9ROSA; Auxin response factor | ||||
| STRING | XP_010095167.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8991 | 32 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21388604 |



