 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010095299.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 484aa MW: 53697.1 Da PI: 5.0991 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.7 | 4.1e-16 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+grWT+eEde+l ++++ G g+W++ ++ g+ R++k+c++rw +yl
XP_010095299.1 14 KGRWTAEEDEILTKYIQANGEGSWRSLPKNAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 57.5 | 3.1e-18 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ + eE+e++v++++ lG++ W++Ia++m+ gRt++++k++w+++l
XP_010095299.1 67 RGNISSEEEEIIVKLHATLGNR-WSLIASHMP-GRTDNEIKNYWNSHL 112
7899******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-21 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.041 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.37E-28 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.0E-12 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.1E-14 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.31E-9 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.861 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-25 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.0E-17 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-15 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.58E-12 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051555 | Biological Process | flavonol biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 484 aa Download sequence Send to blast |
MGRAPCCEKV GLKKGRWTAE EDEILTKYIQ ANGEGSWRSL PKNAGLLRCG KSCRLRWINY 60 LRADLKRGNI SSEEEEIIVK LHATLGNRWS LIASHMPGRT DNEIKNYWNS HLSRKIHIFR 120 RPITSATNTS PTDQTIPKIT DLTKVLNVPP KRKGGRASRL ASMKKKKKKN RSNIVNNNMQ 180 EDHGSSSTVS KNKTNDPKDQ GSSSSANDDH DHGEVVINHE LEIPEPQTPT MEKEITSLSL 240 KALDCDVDDH KDLDDYLLTE PISPPCHEAD HDHDDQSGSG IAQAGSSSDG EKKITSTDQN 300 SSTLLLCPSF TEEEALGPYD DKEGMLCFDD IIDSELLNPS GVLTLSEDSI GSIGAAQNNN 360 TICTSPNKDI NNSGNLSSNG DQICTTESWF SSSSMASSGI NFDDGGVDLW GWENTSKSTV 420 ALGHDHHHHH HEEAMWDQEK EKMLSLLWES DNWEGDHEDD NNHKFGSEII NSDKQNAMVA 480 WLLS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 6e-23 | 12 | 116 | 2 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 2e-22 | 12 | 116 | 56 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 2e-22 | 12 | 116 | 56 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h8a_C | 9e-23 | 12 | 116 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| 1mse_C | 6e-23 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 6e-23 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 150 | 168 | KRKGGRASRLASMKKKKKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
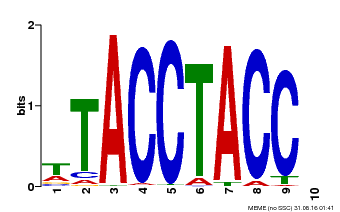
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010095299.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010095299.1 | 0.0 | transcription factor MYB12 | ||||
| TrEMBL | W9QYI9 | 0.0 | W9QYI9_9ROSA; Transcription factor | ||||
| STRING | XP_010095299.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49330.1 | 2e-75 | myb domain protein 111 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21385492 |



