 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010096464.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 335aa MW: 37009.6 Da PI: 6.9767 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 66.8 | 4.3e-21 | 158 | 207 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+W+AeIrd + + +r++lg+f+tae Aa+a+++a+++++g
XP_010096464.1 158 KYRGVRQRP-WGKWAAEIRDpF-----KaARVWLGTFDTAESAARAYDEAALRFRG 207
8********.**********66.....56*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 4.8E-33 | 157 | 216 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 25.541 | 158 | 215 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-23 | 158 | 216 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.2E-14 | 158 | 207 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.6E-36 | 158 | 221 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.78E-36 | 158 | 217 | No hit | No description |
| PRINTS | PR00367 | 6.4E-12 | 159 | 170 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.4E-12 | 181 | 197 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009749 | Biological Process | response to glucose | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 335 aa Download sequence Send to blast |
MNAMVSALSR VVSGNLPKNS YSADHQVSDF SGSVNSSDFL GSNFVGIKRG RDQDEDFRVF 60 GDHQVSSNRV SSSSLLGGDN MQIKLHDLRL TIFISHNIEQ TTEGSNNTWT AATATAMVEN 120 TMSSSTIFPT QTMSFTPTYE YHDMSPQQQQ EQTTRPRKYR GVRQRPWGKW AAEIRDPFKA 180 ARVWLGTFDT AESAARAYDE AALRFRGNKA KLNFPENVRI RPPPVNSPAT HVAVSNSSNT 240 LLSISTTSNP IVHSQSLQSS STTMPLQYSS YNSQFVGSET GTMSLYDQMF SSPPMVDMPF 300 SSSATPESST SNYSDFSEQQ QWTGSSHYNT KNSSG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 1e-24 | 157 | 218 | 1 | 63 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00584 | DAP | Transfer from AT5G64750 | Download |
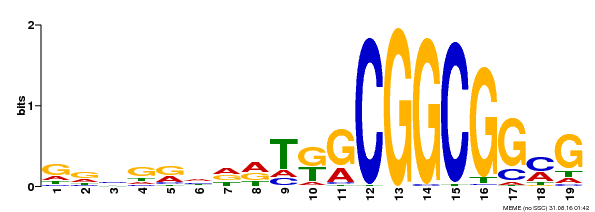
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010096464.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024021449.1 | 0.0 | ethylene-responsive transcription factor ABR1 | ||||
| TrEMBL | W9RA19 | 0.0 | W9RA19_9ROSA; Ethylene-responsive transcription factor | ||||
| STRING | XP_010096464.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4933 | 32 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50080.1 | 7e-29 | ethylene response factor 110 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21390133 |



