 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010099404.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 315aa MW: 34877.8 Da PI: 8.3095 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.7 | 9.3e-19 | 166 | 220 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ ++tk+q Lee F+++++++ ++++ LA++l+L rqV vWFqNrRa+ k
XP_010099404.1 166 RKKLRLTKDQSALLEESFKQHSTLNPKQKQALARQLNLRPRQVEVWFQNRRARTK 220
788899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 125 | 3.4e-40 | 166 | 255 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
+kk+rl+k+q++lLEesF+++++L+p++K++lar+L+l+prqv+vWFqnrRARtk+kq+E+d+e+Lk+++++l++en+rL+ke +eL+ +l
XP_010099404.1 166 RKKLRLTKDQSALLEESFKQHSTLNPKQKQALARQLNLRPRQVEVWFQNRRARTKLKQTEVDCEFLKKCCETLTDENRRLHKELQELK-AL 255
69*************************************************************************************9.55 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.59E-18 | 158 | 223 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.524 | 162 | 222 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.1E-15 | 164 | 226 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 3.6E-16 | 166 | 220 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-17 | 166 | 220 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 6.24E-16 | 166 | 223 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 197 | 220 | IPR017970 | Homeobox, conserved site |
| Pfam | PF02183 | 4.6E-10 | 222 | 256 | IPR003106 | Leucine zipper, homeobox-associated |
| SMART | SM00340 | 1.7E-25 | 222 | 265 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 315 aa Download sequence Send to blast |
MGFDHDHVCN TGLVLRLGFS PTTATATATA AKKPSTTTNS SSTPFEPSLL TLGLSGGGDE 60 AHDNNNNNNN SSSNNNNNRK IIDVSKSFHD HHLHQEPVQN NNNSNNNISV HDNSLYRQAS 120 PHSAVSSFSG GRVKRERDLS SEEIEVEEKV SLSRVSDEDE DGTNARKKLR LTKDQSALLE 180 ESFKQHSTLN PKQKQALARQ LNLRPRQVEV WFQNRRARTK LKQTEVDCEF LKKCCETLTD 240 ENRRLHKELQ ELKALKLAQP LYMHMPAATL TMCPSCERIV GGVGGDVVGG SAKSPFSMAP 300 KPHFYNPFTN PSAAC |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 214 | 222 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00477 | DAP | Transfer from AT4G37790 | Download |
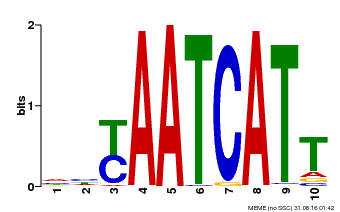
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010099404.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010099404.1 | 0.0 | homeobox-leucine zipper protein HAT22 | ||||
| Swissprot | P46604 | 1e-97 | HAT22_ARATH; Homeobox-leucine zipper protein HAT22 | ||||
| TrEMBL | W9RI15 | 0.0 | W9RI15_9ROSA; Homeobox-leucine zipper protein HAT22 | ||||
| STRING | XP_010099404.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1144 | 33 | 107 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37790.1 | 4e-81 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21391159 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



