 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010101635.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 718aa MW: 78938.2 Da PI: 7.9872 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 37.8 | 3.4e-12 | 457 | 495 | 2 | 44 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiL 44
r +h++ E+rRR++iN++f+ Lrel+P+ ++K +Ka+ L
XP_010101635.1 457 RSKHSATEQRRRSKINDRFQMLRELIPHT----DQKRDKASFL 495
889*************************8....8999999987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.2E-13 | 452 | 496 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 7.98E-11 | 454 | 496 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 12.227 | 455 | 505 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.26E-9 | 457 | 495 | No hit | No description |
| Pfam | PF00010 | 8.9E-10 | 457 | 495 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 718 aa Download sequence Send to blast |
MRIKSFVTSL LSEYDHIVTT ILHGKDKITF DEVCAALYNN EIWKKDQKEH KSTSAAKYED 60 DESEFVLIST LSKSQYDEWI MDSGCTYHMC SNKEWFFNFE ELNEGVVNIG TNDALQSTGI 120 GSIHLKCHDG STKVLTDVRY VPHLKKNHIF LGVLGSKGFT VTMRDGILKV VSSTLVNLVK 180 HGLLKGAKTG KLEFYERCVL AKQLKVKFGR KPTHDFLSLY SHSTTQQDPR PPSQGGYLKT 240 HDFLQPLERV GKTCAKEETS VEISTSEKPP PPAPPPVEHV LPGGIGTYSI SHISNYFNQR 300 VPKPEGMVFT AAQASSTDRN DENSNCSSYT GSGFTLWEES AVKKGKTGKE NMAEKASVLE 360 PAARVGQWAS SERPSQSSSN NHRGSFSSLS SSQPSGHRSR SFMEMIKSAK CSSQDEELED 420 EEPFVLKKET STVTTTHKEL RVKVDGKSSD QKANTPRSKH SATEQRRRSK INDRFQMLRE 480 LIPHTDQKRD KASFLLERNN HRIGESFDHS QGMNGGPGPA LVFTGKFEEK HISVSPTVPG 540 SAQNPVESDT STATITNKGL SFPMSLQPNF FSPLRNGGEV PQAPSSHPSA VENTSPQPQS 600 QLCQTRSCTT VDGAVATDKL KEEELTIEGG TISISSAYSQ GLLNSLTQAL QSSGVDLSQA 660 SISVQIELGK RANSRTAATT SITKDNEFPS SNQEIKQSRL ATGDDSDLAQ KKLKTGKN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Transcription factor that bind specifically to the DNA sequence 5'-CANNTG-3'(E box). Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BZR2/BES1. Does not itself activate transcription but enhances BZR2/BES1-mediated target gene activation. {ECO:0000269|PubMed:15680330}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
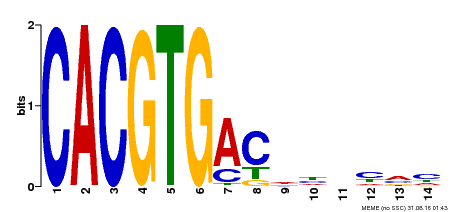
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010101635.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024024837.1 | 0.0 | transcription factor BIM1 isoform X2 | ||||
| Swissprot | Q9LEZ3 | 1e-104 | BIM1_ARATH; Transcription factor BIM1 | ||||
| TrEMBL | W9RRJ1 | 0.0 | W9RRJ1_9ROSA; Uncharacterized protein | ||||
| STRING | XP_010101635.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7252 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.5 | 7e-99 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21399703 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



