 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010102909.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 336aa MW: 38721.6 Da PI: 7.3181 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.7 | 1.4e-18 | 7 | 54 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd l+++++ +G g+W++ ar+ g++Rt+k+c++rw++yl
XP_010102909.1 7 RGPWTVEEDRTLINYIANHGEGRWNSLARCAGLKRTGKSCRLRWLNYL 54
89********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.2 | 2.9e-16 | 60 | 103 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ T eE++l+++++ ++G++ W++Ia++++ gRt++++k++w++
XP_010102909.1 60 RGNITLEEQLLILELHSRWGNR-WSKIAQHLP-GRTDNEIKNYWRT 103
7999******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 24.248 | 2 | 58 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.49E-31 | 4 | 101 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-15 | 6 | 56 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.6E-17 | 7 | 54 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-23 | 8 | 61 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.21E-12 | 9 | 54 | No hit | No description |
| PROSITE profile | PS51294 | 18.707 | 59 | 109 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.3E-14 | 59 | 107 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.8E-15 | 60 | 103 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.4E-23 | 62 | 108 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009620 | Biological Process | response to fungus | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MMMDLRRGPW TVEEDRTLIN YIANHGEGRW NSLARCAGLK RTGKSCRLRW LNYLRPDVRR 60 GNITLEEQLL ILELHSRWGN RWSKIAQHLP GRTDNEIKNY WRTRVQKHAK QLKCDVNSKQ 120 FKDTMRYLWM PRLVERIQAA NTAAATTTAN ITVSAGAPST HHHQFNNFHN NNMINNNLPI 180 SNIINISNDF VGSANYTPEN SSTAASSDSF GAQFSPVSDL TDHYFTANNN PTATTTTLPV 240 NNNSNTTTTT TNPHDHHQYF QAAPSTTHDH HQQVGGTYYY HPSMSTTYDQ NNMSFSHEGL 300 NIDDRFQASM DQNNQYWIEE VDNLWNNVDQ DMWFLQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 5e-25 | 4 | 101 | 24 | 120 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00334 | DAP | Transfer from AT3G06490 | Download |
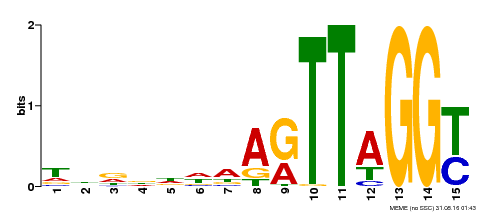
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010102909.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010102909.2 | 0.0 | transcription factor MYB108 | ||||
| TrEMBL | W9RJC4 | 0.0 | W9RJC4_9ROSA; Myb-related protein 305 | ||||
| STRING | XP_010102909.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF875 | 34 | 125 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G06490.1 | 8e-88 | myb domain protein 108 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21389421 |



