 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010103252.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 463aa MW: 52870.4 Da PI: 8.3237 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 67.5 | 1.8e-21 | 118 | 178 | 2 | 57 |
T--SS--HHHHHHHHHHHHH.SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 2 rkRttftkeqleeLeelFek.nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
r+R+t+t+ ql++Le++Fe+ n +ps+++++e+++ l +++e++V++WFqNrRa+ k+
XP_010103252.1 118 RQRWTPTPVQLQILERMFEQgNGTPSKQKIKEITSDLsqhgQISETNVYNWFQNRRARSKR 178
89*****************99**************************************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 99.7 | 2.1e-32 | 117 | 180 | 2 | 65 |
Wus_type_Homeobox 2 artRWtPtpeQikiLeelyksGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
ar+RWtPtp Q++iLe+++++G tP+k++i++it+ L+++G+i+++NV++WFQNr+aR+++kq
XP_010103252.1 117 ARQRWTPTPVQLQILERMFEQGNGTPSKQKIKEITSDLSQHGQISETNVYNWFQNRRARSKRKQ 180
789***********************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 7.8E-18 | 113 | 183 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 15.224 | 114 | 179 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.1E-15 | 116 | 183 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.52E-18 | 117 | 181 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.60E-13 | 118 | 180 | No hit | No description |
| Pfam | PF00046 | 2.9E-19 | 118 | 178 | IPR001356 | Homeobox domain |
| Pfam | PF10181 | 1.2E-15 | 361 | 424 | IPR019328 | GPI-GlcNAc transferase complex, PIG-H component, conserved domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0017176 | Molecular Function | phosphatidylinositol N-acetylglucosaminyltransferase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 463 aa Download sequence Send to blast |
MGWEKQQLQL QLQLQLQQQQ QTQEDQNMRE RSEQQQQVNN NSNNNLAFHH HHHHVKVMTD 60 EQLETLRKQI SVYATICEQL VQMHKNLTSH QDLPPGVRLG NLYCDPLMTS AGHKITARQR 120 WTPTPVQLQI LERMFEQGNG TPSKQKIKEI TSDLSQHGQI SETNVYNWFQ NRRARSKRKQ 180 HNSTPNNAES EVETEVESPK DKKTKPEELL SQHNSAPRAE DICFQNPEIS SDLHFFDPQS 240 NKGEAIFTSN TCLRPPARNL AQMPFYDGLL SISNSRMVDS RYSYSYSYSG EGKGSVDIHI 300 VARKNGVKGG FFVFFSALLI LANFFYLFQL KGNSIVIVLW SFVIEALLLK LLLWKAVKKE 360 SVLVMPAFGV QLETHYVSGK VIRRFIPVDK ILKPVVLECV TPVTCYWCLC LVVCGEAELI 420 SVFETLRPPV KILVPIWKAL RAATNKEEIP NTSITENGEG GLQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 171 | 179 | RRARSKRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00617 | PBM | Transfer from AT4G35550 | Download |
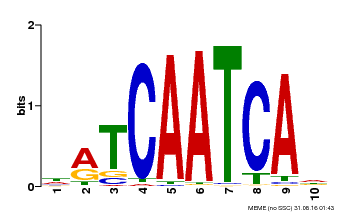
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010103252.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024025789.1 | 0.0 | WUSCHEL-related homeobox 8 isoform X1 | ||||
| TrEMBL | W9S9Q3 | 0.0 | W9S9Q3_9ROSA; WUSCHEL-related homeobox 13 | ||||
| STRING | XP_010103252.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1855 | 34 | 93 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G35550.1 | 5e-83 | WUSCHEL related homeobox 13 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21390482 |



