 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010104441.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 406aa MW: 45007.6 Da PI: 8.4467 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 82.4 | 3.8e-26 | 143 | 208 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
r+++sL+llt+kf+kl+ ++e+++++ln+ a L +v ++RRiYDi+NVLe+++liek++kn+irwkg
XP_010104441.1 143 RYDSSLGLLTKKFVKLILEAEDRTLDLNHTAGVL---EV--QKRRIYDITNVLEGVGLIEKTSKNHIRWKG 208
6899******************************...99..****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 1.7E-27 | 141 | 209 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 2.5E-34 | 143 | 208 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.1E-16 | 143 | 206 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF02319 | 1.2E-23 | 145 | 208 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| CDD | cd14660 | 1.97E-36 | 219 | 326 | No hit | No description |
| SuperFamily | SSF144074 | 8.89E-26 | 220 | 325 | No hit | No description |
| Pfam | PF16421 | 6.3E-23 | 224 | 323 | IPR032198 | E2F transcription factor, CC-MB domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000902 | Biological Process | cell morphogenesis | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042023 | Biological Process | DNA endoreduplication | ||||
| GO:0051782 | Biological Process | negative regulation of cell division | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 406 aa Download sequence Send to blast |
MMDDPSTAHG FQFQLLHSHP HPLSHHHSSS SPSNPHLFPS SSSSSSSPLR PHFVAPPSNH 60 SHHRLCDFSK NSQLVQSNVK YEAQIWSPAV AGQSKVNDPA AERKPSGLGT YNIKSKVPKQ 120 TKIGSQRLKV ECLNGLSPSI NCRYDSSLGL LTKKFVKLIL EAEDRTLDLN HTAGVLEVQK 180 RRIYDITNVL EGVGLIEKTS KNHIRWKGYD GLGPSKLIDE IAGLKAEVGS LYAEEYRLDE 240 AIRDKQDRLR ALGEDENCKK YLFLTVDDIL TLPCFQNQTL IAIKAPTASY IEVPDPDADI 300 SFSQRQYKLI VRSSRGPIDL YLLSKYQGAH EEATTKQSSK KSSSVRDGEG LSFDHQGNQK 360 VSSFSLGSLG SKASGIQKII PANVDANDDY WFRSDPEVSI TNLWGK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1cf7_A | 2e-21 | 138 | 214 | 2 | 76 | PROTEIN (TRANSCRIPTION FACTOR E2F-4) |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00190 | DAP | Transfer from AT1G47870 | Download |
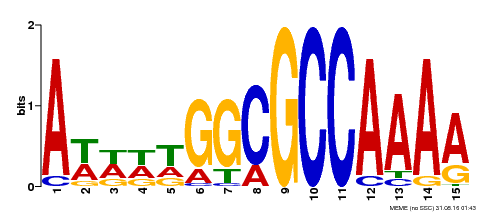
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010104441.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024026586.1 | 0.0 | transcription factor E2FC isoform X2 | ||||
| TrEMBL | W9SD41 | 0.0 | W9SD41_9ROSA; Transcription factor E2FC | ||||
| STRING | XP_010104441.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11885 | 25 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G47870.1 | 1e-101 | E2F/DP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21399112 |



