 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010104815.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 349aa MW: 39476.2 Da PI: 5.5436 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 64.8 | 1.7e-20 | 198 | 248 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpseng.krkrfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+++AeIrdp + + r++lg+f+ta eAaka+++a+ kl+g
XP_010104815.1 198 HYRGVRQRP-WGKYAAEIRDP---NrRGSRVWLGTFDTAIEAAKAYDRAAFKLRG 248
7********.**********9...4334*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 1.8E-32 | 197 | 257 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.25 | 198 | 256 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.9E-36 | 198 | 262 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.84E-31 | 198 | 256 | No hit | No description |
| Pfam | PF00847 | 2.0E-14 | 198 | 248 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 8.5E-23 | 198 | 257 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 1.4E-13 | 199 | 210 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.4E-13 | 238 | 258 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 349 aa Download sequence Send to blast |
MATINEVSKL ELIKQYLLGD LPPVSSFSNF DRNQWISDLK TEFSSSQSQS DSHCSRTSSS 60 SDYLDSSDVL DFTSDFEPKP DNFDFDFDFD FSEFEMKPEL IDLTTPKSRN LTSQSSDASF 120 EFESKPQIID LRSHIPQSSS ISFEFESKPQ ISGQKGSQLN RKPSLTISLP AKTEWIQFSA 180 PVPPASVQKP AVEEEIRHYR GVRQRPWGKY AAEIRDPNRR GSRVWLGTFD TAIEAAKAYD 240 RAAFKLRGSK AILNFPLEAG KYDVVVRTDK KKRRRQEEGG YVSEVVKEPK KEKVTEFLDT 300 VSYVKNMPLT PSNWTAVWDS SDGQGVFNVP PLSPLSPHPS MGYPQLMVV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 2e-29 | 197 | 257 | 1 | 61 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| 2gcc_A | 2e-29 | 197 | 257 | 4 | 64 | ATERF1 |
| 3gcc_A | 2e-29 | 197 | 257 | 4 | 64 | ATERF1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 269 | 274 | KKKRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00549 | DAP | Transfer from AT5G47230 | Download |
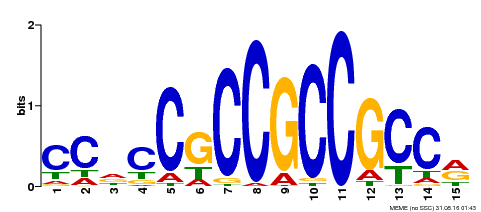
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010104815.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010104815.1 | 0.0 | ethylene-responsive transcription factor 5 | ||||
| TrEMBL | W9RPJ4 | 0.0 | W9RPJ4_9ROSA; Ethylene-responsive transcription factor 5 | ||||
| STRING | XP_010104815.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1085 | 34 | 109 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47230.1 | 2e-58 | ethylene responsive element binding factor 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21406765 |



