|
Plant Transcription
Factor Database
|
Transcription Factor Information
|
Basic
Information? help
Back to Top |
| TF ID |
XP_010104868.1 |
| Organism |
|
| Taxonomic ID |
|
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
| Family |
MYB_related |
| Protein Properties |
Length: 293aa MW: 31493.7 Da PI: 5.1515 |
| Description |
MYB_related family protein |
| Gene Model |
| Gene Model ID |
Type |
Source |
Coding Sequence |
| XP_010104868.1 | genome | BGI | View CDS |
|
| Signature Domain? help Back to Top |
 |
| No. |
Domain |
Score |
E-value |
Start |
End |
HMM Start |
HMM End |
| 1 | Myb_DNA-binding | 38.7 | 2.3e-12 | 57 | 101 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ ++ + ++lG+g+W+ Iar Rt+ q+ s+ qky
XP_010104868.1 57 PWTEEEHRMFLLGLQKLGKGDWRGIARNYVISRTPTQVASHAQKY 101
8*****************************89************9 PP
|
| Gene Ontology ? help Back to Top |
| GO Term |
GO Category |
GO Description |
| GO:0000122 | Biological Process | negative regulation of transcription from RNA polymerase II promoter |
| GO:0009651 | Biological Process | response to salt stress |
| GO:0009723 | Biological Process | response to ethylene |
| GO:0009737 | Biological Process | response to abscisic acid |
| GO:0009739 | Biological Process | response to gibberellin |
| GO:0009751 | Biological Process | response to salicylic acid |
| GO:0009753 | Biological Process | response to jasmonic acid |
| GO:0030307 | Biological Process | positive regulation of cell growth |
| GO:0046686 | Biological Process | response to cadmium ion |
| GO:0048366 | Biological Process | leaf development |
| GO:2000469 | Biological Process | negative regulation of peroxidase activity |
| GO:0005634 | Cellular Component | nucleus |
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding |
| Sequence ? help Back to Top |
| Protein Sequence Length: 293 aa
Download sequence Send
to blast |
MGTLSHYAGS GSTPGGPNVP GSPGETPDHG GAADGYASED FVPGSSSSSR ERKKGVPWTE 60
EEHRMFLLGL QKLGKGDWRG IARNYVISRT PTQVASHAQK YFIRQTNVSR RKRRSSLFDI 120
VADESADAPM VPRDFLSVNQ PQMELPNENT SLDPPAIDEE CESMDSTNSN DGEPVPLNPD 180
SSQTCYPIIY PAYVSPFFPF PIPFWSGYDN GTEQNKETHE VVKPTAVHSK SPINVDELVG 240
MSKLSLGDSI GNAGPSPLSL KLLEGSSRQS AFHANPGSSS SNMNSSGSPI HAV
|
| Functional Description ? help
Back to Top |
| Source |
Description |
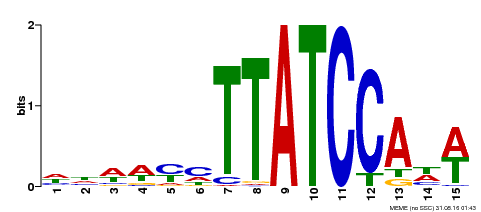
| UniProt | Transcriptional repressor (PubMed:23888064, PubMed:24806884). Direct regulator of the transcription of peroxidase (Prxs) and reactive oxygen species (ROS)-related genes via the recognition of 5'-ATCACA-3' motif (PubMed:24806884). Binds to 5'-TATCCA-3' motif (TA box) and represses the activity of corresponding promoters (e.g. sugar response genes) (PubMed:25920996). Regulates hypocotyl elongation in response to darkness by enhancing auxin accumulation in a phytochrome-interacting factor (PIF) proteins-dependent manner. Promotes lateral roots formation (PubMed:23888064). Promotes cell expansion during leaves development via the modulation of cell wall-located Prxs (PubMed:24806884). Plays a critical role in developmentally regulated and dark-induced onset of leaf senescence by repressing the transcription of several genes involved in chloroplast function and responses to light and auxin. Promotes responses to auxin, abscisic acid (ABA), and ethylene (PubMed:25920996). {ECO:0000269|PubMed:23888064, ECO:0000269|PubMed:24806884, ECO:0000269|PubMed:25920996}. |
| Regulation -- Description ? help
Back to Top |
| Source |
Description |
| UniProt | INDUCTION: Slightly induced by CdCl(2) (PubMed:16463103). Accumulates in the dark (PubMed:23888064, PubMed:25920996). Diurnal expression pattern with maximal levels in the morning (at protein level). Specifically induced during leaf expansion (PubMed:24806884). Expressed in old and dark-treated leaves (PubMed:25920996). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:23888064, ECO:0000269|PubMed:24806884, ECO:0000269|PubMed:25920996}. |