 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010105426.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 422aa MW: 47650.4 Da PI: 6.9715 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.6 | 1e-31 | 90 | 143 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH +Fv+ave+LGG+e+AtPk +l++m++kgL+++hvkSHLQ+YR+
XP_010105426.1 90 PRLRWTPDLHLCFVRAVERLGGQERATPKLVLQMMNIKGLSIAHVKSHLQMYRS 143
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.142 | 86 | 146 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-29 | 86 | 144 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.02E-14 | 87 | 144 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-22 | 90 | 145 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.7E-9 | 91 | 142 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 422 aa Download sequence Send to blast |
MEKSIDSGSK VISKTSPSNN YNINNEYQDD EENEEEEEEE DQEKADDSGQ QNINGRSSSN 60 STVEENNNGS SKKGSASGGS VRQYIRSKTP RLRWTPDLHL CFVRAVERLG GQERATPKLV 120 LQMMNIKGLS IAHVKSHLQM YRSKKIEDPN QDPHSFKTVL SEQGFFLEGG DHQIYNLSQL 180 PMLQGFNQWP TSSGLIRYAD SSWRSSTRDH QIYSPQYSSF RTAFDSAKNG VYGSVAERLF 240 RNNNNSINIG SATSTLDTSA SWRSRHQTHD QGESFRRSWR TQIFNNPSST ELDLSLQNKR 300 IMLHEKGFMR DHQVVNTITT YNLSTSSTNS RSVQEGGQNK LKRKVLDSER DSDLDLNLSL 360 KIRPKDDHDH EEVDINSTSL CLSLSPSSSS NKLIMSRLKE EAGGLIQDRK LGRTASTLDL 420 TL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 1e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 1e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 1e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 1e-17 | 91 | 145 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-17 | 91 | 145 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
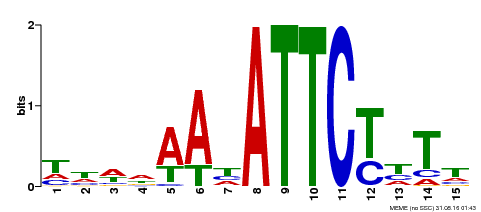
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010105426.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015877428.1 | 1e-114 | putative Myb family transcription factor At1g14600 | ||||
| TrEMBL | W9SFW5 | 0.0 | W9SFW5_9ROSA; Putative Myb family transcription factor | ||||
| STRING | XP_010105426.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2378 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 8e-49 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21393209 |



