 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010105670.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 844aa MW: 93201.4 Da PI: 6.1059 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.1 | 6.8e-35 | 135 | 211 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cqv+ C ad+ e+k yhrrh+vC +++a +v+++g+++r+CqqC++fh s+fDe+krsCrr+L++hn+rrr+k
XP_010105670.1 135 RCQVPTCGADIRELKGYHRRHRVCLRCANAGTVVIEGVNKRYCQQCGKFHVSSDFDEGKRSCRRKLERHNNRRRRKP 211
6************************************************************************9975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.2E-26 | 130 | 197 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 28.65 | 133 | 210 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.96E-32 | 134 | 213 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.9E-25 | 136 | 210 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 844 aa Download sequence Send to blast |
MEGASSPSSD HARDSEMEVG HHAPSVHDDA PPNVWDWGDL LDFAVDADLS INWDSGPAEP 60 PPPMEVPAES PVREVLDPGR IRKRDPRMVC SNFLAGRVPC ACPEMDEKMM EMEEDEAGHG 120 KKRARTARAQ PGAARCQVPT CGADIRELKG YHRRHRVCLR CANAGTVVIE GVNKRYCQQC 180 GKFHVSSDFD EGKRSCRRKL ERHNNRRRRK PVDSKGAIEK ESQGDAQSED ASGDGPDGEG 240 GKDCSQFSSQ MVQKETWVDS EAGHASPLSP LRTAPDSKDA NSDGFELINS GETQVDGGKH 300 NSRRGLSPSY YENKSAYSSV CPTGRISFKL YDWNPAEFPR RLRHQIFQWL ANMPVELEGY 360 IRPGCIILTV FVAMPRFMWM KLSEDPVSYI HNFVVAPGGM LSGRGNILVY VNNMVFQVVK 420 GGNSVIKAKV DVGVPRLHYV HPTCFEAGKP MEFVACGSNL FQPKLRFLLS FSGKYLAYDY 480 SSASSRFQTA SNLDHQLYRI QVPHTEADCF GPVFIEVENE AGLSNFIPVL IGDKETCSEM 540 KVIQQRLDES LLKDGPCVSP IASLSNSCDA SSLRQSAITE LILDVAWLLK KPGSEGFQQI 600 LTASQVQRLN RLLSLLISVE STTILERILQ NMKSVMDKLK LTDECSGISD ADLRLLQKYM 660 DYAHQLSYQK LQKDGSSDLP SRNLPLKEDQ CCCPDDACSD VSFLCQDTKI MVNGKLGLVA 720 SSTYSKTSET IRLLTNETFS EANLIKEWPS DASCHTASGE LMSSRNNSVP TTSDIDKLPR 780 GFNHVISALS IATERINFLE AVIAASSKCI SCTAVLDVSW DVRSLPLPNS DLPTALRLFS 840 GSDA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 6e-40 | 134 | 216 | 4 | 86 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 196 | 208 | RRKLERHNNRRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
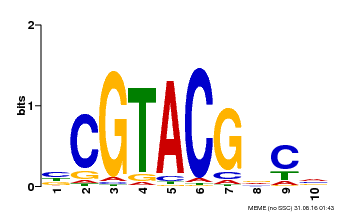
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010105670.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024027509.1 | 0.0 | squamosa promoter-binding-like protein 7 | ||||
| TrEMBL | W9SGM9 | 0.0 | W9SGM9_9ROSA; Squamosa promoter-binding-like protein 7 | ||||
| STRING | XP_010105670.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6470 | 34 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.3 | 0.0 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21394545 |



