 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010105895.1 | ||||||||
| Common Name | DREB6B | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 354aa MW: 38720.2 Da PI: 7.6804 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65 | 1.5e-20 | 166 | 215 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+WvAeIr p++ r+r +lg+f+taeeAa a+++a+ kl+g
XP_010105895.1 166 LYRGVRQRH-WGKWVAEIRLPKN---RTRLWLGTFDTAEEAALAYDRAAYKLRG 215
59******9.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 2.58E-34 | 165 | 225 | No hit | No description |
| SMART | SM00380 | 7.6E-38 | 166 | 229 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.7E-23 | 166 | 224 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.117 | 166 | 223 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.0E-32 | 166 | 224 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.4E-14 | 167 | 215 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.4E-11 | 167 | 178 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.4E-11 | 189 | 205 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0045595 | Biological Process | regulation of cell differentiation | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MAATMDFYSS RPLQSELMEA LEPFMRSASP SSSTITTLSP SSSSPSPPST SSPSYNYFSF 60 SPSIQAQAQT GYGCSTSTAQ MFSDGFSNQA LLGFEQQPGS IGLNQLTPSQ IHQIQAQICQ 120 IQAQTQPNLA YPTHNHHQQQ LHSLSFLGPR PVPMKQVGSP PKPTKLYRGV RQRHWGKWVA 180 EIRLPKNRTR LWLGTFDTAE EAALAYDRAA YKLRGDFARL NFPHLRHHGS SIGGGEFGDY 240 KPLHSSVDAK LQAICESLAE SQSQQKQGVK KKPPAKRSSS AKQEVHEPKV EISPSPPSPV 300 VTTESDGSAG SSPLSDLTFP DIVESPWDCN NGGLESNYLL EKYPSEIDWA SILA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 6e-22 | 165 | 223 | 1 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
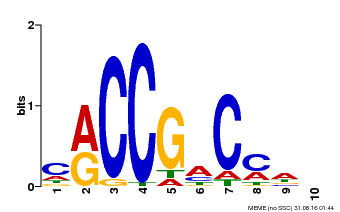
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00026 | PBM | Transfer from AT1G78080 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010105895.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF678396 | 0.0 | KF678396.1 Morus notabilis dehydration responsive element binding transcription factor (DREB6B) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010105895.1 | 0.0 | ethylene-responsive transcription factor RAP2-4 | ||||
| Swissprot | Q8H1E4 | 2e-90 | RAP24_ARATH; Ethylene-responsive transcription factor RAP2-4 | ||||
| TrEMBL | W6FFW5 | 0.0 | W6FFW5_9ROSA; Dehydration responsive element binding transcription factor | ||||
| STRING | XP_010105895.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1057 | 32 | 109 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78080.1 | 8e-59 | related to AP2 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21404406 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



