 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010108641.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Moraceae; Morus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 611aa MW: 68941 Da PI: 6.5192 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 96.3 | 2.8e-30 | 82 | 166 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW++qe+laL+++r++m+ ++r+++lk+plWeevs+k++e g++rs k+Ckek+en+ k+++++k+g+ +++ + +t+++fdqlea
XP_010108641.1 82 RWPRQETLALLKIRSDMDATFRDSSLKAPLWEEVSRKLAELGYKRSGKKCKEKFENVYKYHRRTKDGRTGKS--DGKTYRFFDQLEA 166
8********************************************************************964..5557*******85 PP
| |||||||
| 2 | trihelix | 101.8 | 5.3e-32 | 428 | 513 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW+k ev aLi++r++++ +++++ k+plWe++s+ mr+ g+ rs+k+Ckekwen+nk++kk+k+++kkr s++s+tcpyf+ql+a
XP_010108641.1 428 RWPKAEVEALINLRTNLDLKYEENVPKGPLWEDISAGMRRLGYDRSSKRCKEKWENINKYFKKVKDSDKKR-SDDSKTCPYFHQLDA 513
8*********************************************************************8.89999********85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 7.654 | 75 | 139 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.0016 | 79 | 141 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 1.1E-19 | 80 | 166 | No hit | No description |
| CDD | cd12203 | 2.00E-24 | 81 | 146 | No hit | No description |
| SMART | SM00717 | 1.2 | 425 | 487 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 5.9E-22 | 427 | 514 | No hit | No description |
| PROSITE profile | PS50090 | 7.271 | 427 | 485 | IPR017877 | Myb-like domain |
| CDD | cd12203 | 5.42E-23 | 430 | 492 | No hit | No description |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 611 aa Download sequence Send to blast |
MGDASTTATG SAFGSSGEAA AAAAAEEDRH EEVLLRHEEN GDGDGNGAGS AFLVNNSAEE 60 DDKEDRNVED GGDRSDNFGR NRWPRQETLA LLKIRSDMDA TFRDSSLKAP LWEEVSRKLA 120 ELGYKRSGKK CKEKFENVYK YHRRTKDGRT GKSDGKTYRF FDQLEAFDQN HHPPPPPLQA 180 AAKVPHQAIT NPPISTNVSH NIFVNPNPTS NNISITLPPP PPPPTSTTSN SLPSPQTIIT 240 KSMITNHNNN INGLLFSSPT SSSTASDEEF QARHKRKRKW RDFFRRLTRE VVRKQEEMHK 300 RFLETVAKCE HERTAREEAW RLQEMARINR EHEILLQERS AATAKDAALI AFLQKVSGVT 360 QNLQIISTDN VALSVLNNSI NNNHHNNNTN NHTISLPAPQ PAAKQAATAA PPHVRSLSTT 420 SSSSWSSRWP KAEVEALINL RTNLDLKYEE NVPKGPLWED ISAGMRRLGY DRSSKRCKEK 480 WENINKYFKK VKDSDKKRSD DSKTCPYFHQ LDAIYREKLK NIDPKAQENG APHPQQPPPH 540 FGMEPLMVQP ERQWPLQEEE YDQEEEVDRD TSSSTEVEDQ DHDDHDNHRE GTGDGSYGIV 600 SNVQSSMDMV E |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 124 | 132 | KRSGKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00244 | DAP | Transfer from AT1G76890 | Download |
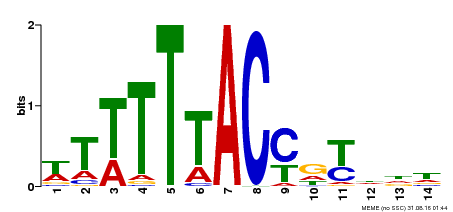
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010108641.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK107139 | 3e-35 | AK107139.1 Oryza sativa Japonica Group cDNA clone:002-124-D01, full insert sequence. | |||
| GenBank | AL606587 | 3e-35 | AL606587.3 Oryza sativa genomic DNA, chromosome 4, BAC clone: OSJNBa0011L07, complete sequence. | |||
| GenBank | AP014960 | 3e-35 | AP014960.1 Oryza sativa Japonica Group DNA, chromosome 4, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012612 | 3e-35 | CP012612.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 4 sequence. | |||
| GenBank | CR855197 | 3e-35 | CR855197.1 Oryza sativa genomic DNA, chromosome 4, BAC clone: H0115B09, complete sequence. | |||
| GenBank | X68261 | 3e-35 | X68261.1 O.sativa gt-2 gene. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010108641.1 | 0.0 | trihelix transcription factor GT-2 | ||||
| Swissprot | Q39117 | 1e-137 | TGT2_ARATH; Trihelix transcription factor GT-2 | ||||
| TrEMBL | W9S035 | 0.0 | W9S035_9ROSA; Trihelix transcription factor GT-2 | ||||
| STRING | XP_010108641.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF373 | 34 | 181 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 1e-70 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 21400857 |



