 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010522381.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 524aa MW: 57853.4 Da PI: 9.0261 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 53.6 | 4.1e-17 | 269 | 318 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lr+l+P++ + K +Ka+ L +++eYI+ Lq
XP_010522381.1 269 RSKHSATEQRRRSKINDRFQMLRQLIPNS----DHKRDKASFLLEVIEYIQFLQ 318
889*************************9....89******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.8E-23 | 264 | 337 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.66E-19 | 265 | 337 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.888 | 267 | 317 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.17E-15 | 269 | 321 | No hit | No description |
| Pfam | PF00010 | 1.2E-14 | 269 | 318 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.2E-13 | 273 | 323 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 524 aa Download sequence Send to blast |
MELPQPRPFE TEGRKPTHDF LSLCSHSTVP PDPRTSSQGG HLKTHDFLQP LERVGRSGTK 60 EETSSSIQTL ASEKPPPPAP PPSSVDQHVL PGGIGTYMIS HIPYFHHRIP KPEVSPVFTV 120 AQASGSERNT DENSNCSSYA PGGFTLWEES ARKKGQTRKE NVGERASMLD AASAGQWPGP 180 ERQSQSSTTT NNRSNFSSRS SSQVSGLKSQ SFMDMIRSAK GSSQDNDGDD DEEEFIMKKE 240 SPSTSQKVDL RVRVDGKSTG NDQKPNTPRS KHSATEQRRR SKINDRFQML RQLIPNSDHK 300 RDKASFLLEV IEYIQFLQEK VNKYEGCYQG WNPEPGKLMN WRNNSQHPAE GTVAFGPPKL 360 EEKNSVIAMT QTAMDQQCPF PMSIKHSSFY AAGPTVLTGS PVSQFQTRVV SEERPKSGSS 420 WEVVASESGK PKGEEGDLVI GGGTISISTV YSQGLMKRLK EALKSSGVDM SKASISVQIE 480 LAKQRPAPST LMDEEVGGKR PRTETGEEEG HSNRNRKKLR PDRI |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 513 | 519 | RNRKKLR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Transcription factor that bind specifically to the DNA sequence 5'-CANNTG-3'(E box). Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BZR2/BES1. Does not itself activate transcription but enhances BZR2/BES1-mediated target gene activation. {ECO:0000269|PubMed:15680330}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
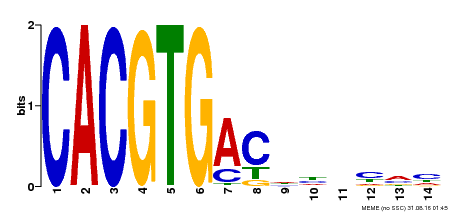
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010522381.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010522381.1 | 0.0 | PREDICTED: transcription factor BIM1-like isoform X1 | ||||
| Swissprot | Q9LEZ3 | 1e-171 | BIM1_ARATH; Transcription factor BIM1 | ||||
| TrEMBL | V4LLR0 | 0.0 | V4LLR0_EUTSA; Uncharacterized protein | ||||
| STRING | XP_010522381.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6967 | 27 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 1e-168 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



