 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010523041.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 815aa MW: 88094.9 Da PI: 6.281 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.1 | 4.8e-21 | 121 | 176 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
XP_010523041.1 121 KKRYHRHTPQQIQELEAMFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 176
688999***********************************************999 PP
| |||||||
| 2 | START | 201.2 | 4.4e-63 | 320 | 544 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetl 82
ela++a++elvk+a+ +ep+Wvk e++n++e++++f+++ + + +ea+r++g+v+ ++ lve+l+d++ +W+e+++ +a+t+
XP_010523041.1 320 ELALTAMDELVKLAQTDEPLWVKGLdderEILNQEEYVRSFPPTVGlkpagFVTEASRTTGMVIINSLALVETLMDSN-RWTEMFPciisRASTT 413
5899*******************999999*************9877********************************.**************** PP
EEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-E CS
START 83 evissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehv 170
+vis g ga+qlm aelq+lsplvp R+ f+R+++q+ +g+w++vdvSvd ++++ + + ++lpSg+++++++ng+skvtwveh+
XP_010523041.1 414 DVISGGmagtrnGAIQLMHAELQVLSPLVPvREANFLRFCKQHAEGVWAVVDVSVDGVRENSGCLPAMSLRRLPSGCVVQDMPNGYSKVTWVEHA 508
**************************************************************9555555569*********************** PP
E--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 171 dlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+++++++h+l+r++++sgl +ga++wvatlqrqce+
XP_010523041.1 509 EYEENQVHQLYRPMLRSGLGFGAQRWVATLQRQCEC 544
**********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.41E-20 | 107 | 178 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-22 | 110 | 178 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.265 | 118 | 178 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.3E-18 | 119 | 182 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.01E-18 | 120 | 178 | No hit | No description |
| Pfam | PF00046 | 1.5E-18 | 121 | 176 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 153 | 176 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 41.339 | 311 | 547 | IPR002913 | START domain |
| CDD | cd08875 | 2.70E-122 | 315 | 543 | No hit | No description |
| SuperFamily | SSF55961 | 5.57E-31 | 315 | 544 | No hit | No description |
| SMART | SM00234 | 1.4E-42 | 320 | 544 | IPR002913 | START domain |
| Pfam | PF01852 | 2.0E-55 | 320 | 544 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 9.07E-24 | 565 | 800 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 815 aa Download sequence Send to blast |
MNFGGLLDNS SGGGGGSRIL SDISYVNHPN AAAAADPRGA MSLFSPPFSK SVFSSPGLSL 60 AIVQPERENR GEASMRASGD NLEGSANRRS REEEHESRSG SDNVEGISGD DQDAADKPPR 120 KKRYHRHTPQ QIQELEAMFK ECPHPDEKQR LELSKRLCLE TRQVKFWFQN RRTQMKTQLE 180 RHENALLRQE NDKLRAENMS IREAMRNPIC SSCGGPAMLG DVSLEEQHLR IENARLKDEL 240 DRVCALAGKF LGRSISGDPP LPNSSLELGV GTTASANAGG FGFPPDFGIS STAVRCLPTI 300 PPQPLVSGVT GIDQRSVLLE LALTAMDELV KLAQTDEPLW VKGLDDEREI LNQEEYVRSF 360 PPTVGLKPAG FVTEASRTTG MVIINSLALV ETLMDSNRWT EMFPCIISRA STTDVISGGM 420 AGTRNGAIQL MHAELQVLSP LVPVREANFL RFCKQHAEGV WAVVDVSVDG VRENSGCLPA 480 MSLRRLPSGC VVQDMPNGYS KVTWVEHAEY EENQVHQLYR PMLRSGLGFG AQRWVATLQR 540 QCECLAILMS SSVNPPDNGA ITAGGRKSML KLAQRMTYNF CSGISAPSVQ SWSKLTVGHV 600 GPDVRVMTRK SVDDPGEPPG IVLSAATSVW VPGSPQRLFD FLRDERLRCE WDILSNGGPM 660 QEMAHIAKGQ DEGNSVCLLR SNAMNASQSS MLILQETRID ASGALVVYAP VDIPAMHVVM 720 NGGDSSYVAL LPSGFAILPD GGGPSSPHGG GPASPHGGGV SPRPPAPRGS LLTVAFQILV 780 NSLPTAKLTV ESVETVNNLI TCTVQKIRAA LHSDS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
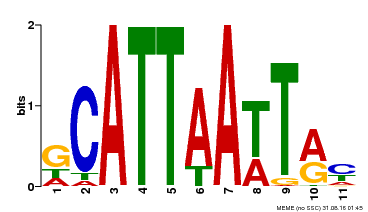
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010523041.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010523041.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X1 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A178V6U4 | 0.0 | A0A178V6U4_ARATH; ANL2 | ||||
| STRING | XP_010523042.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



