 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010523042.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 816aa MW: 88264.1 Da PI: 6.4662 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.1 | 4.8e-21 | 117 | 172 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
XP_010523042.1 117 KKRYHRHTPQQIQELEAMFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 172
688999***********************************************999 PP
| |||||||
| 2 | START | 201.2 | 4.4e-63 | 316 | 540 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetl 82
ela++a++elvk+a+ +ep+Wvk e++n++e++++f+++ + + +ea+r++g+v+ ++ lve+l+d++ +W+e+++ +a+t+
XP_010523042.1 316 ELALTAMDELVKLAQTDEPLWVKGLdderEILNQEEYVRSFPPTVGlkpagFVTEASRTTGMVIINSLALVETLMDSN-RWTEMFPciisRASTT 409
5899*******************999999*************9877********************************.**************** PP
EEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-E CS
START 83 evissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehv 170
+vis g ga+qlm aelq+lsplvp R+ f+R+++q+ +g+w++vdvSvd ++++ + + ++lpSg+++++++ng+skvtwveh+
XP_010523042.1 410 DVISGGmagtrnGAIQLMHAELQVLSPLVPvREANFLRFCKQHAEGVWAVVDVSVDGVRENSGCLPAMSLRRLPSGCVVQDMPNGYSKVTWVEHA 504
**************************************************************9555555569*********************** PP
E--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 171 dlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+++++++h+l+r++++sgl +ga++wvatlqrqce+
XP_010523042.1 505 EYEENQVHQLYRPMLRSGLGFGAQRWVATLQRQCEC 540
**********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.41E-20 | 103 | 174 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-22 | 106 | 174 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.265 | 114 | 174 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.3E-18 | 115 | 178 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.01E-18 | 116 | 174 | No hit | No description |
| Pfam | PF00046 | 1.5E-18 | 117 | 172 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 149 | 172 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 41.339 | 307 | 543 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.57E-31 | 311 | 540 | No hit | No description |
| CDD | cd08875 | 2.52E-122 | 311 | 539 | No hit | No description |
| Pfam | PF01852 | 2.0E-55 | 316 | 540 | IPR002913 | START domain |
| SMART | SM00234 | 1.4E-42 | 316 | 540 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.28E-23 | 561 | 739 | No hit | No description |
| SuperFamily | SSF55961 | 1.28E-23 | 770 | 801 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 816 aa Download sequence Send to blast |
MNFGGLLDNS SGGGGGSRIL SDISYVNHPN ANPRGAMSLF SPPFSKSVFS SPGLSLAIVQ 60 PERENRGEAS MRASGDNLEG SANRRSREEE HESRSGSDNV EGISGDDQDA ADKPPRKKRY 120 HRHTPQQIQE LEAMFKECPH PDEKQRLELS KRLCLETRQV KFWFQNRRTQ MKTQLERHEN 180 ALLRQENDKL RAENMSIREA MRNPICSSCG GPAMLGDVSL EEQHLRIENA RLKDELDRVC 240 ALAGKFLGRS ISGDPPLPNS SLELGVGTTA SANAGGFGFP PDFGISSTAV RCLPTIPPQP 300 LVSGVTGIDQ RSVLLELALT AMDELVKLAQ TDEPLWVKGL DDEREILNQE EYVRSFPPTV 360 GLKPAGFVTE ASRTTGMVII NSLALVETLM DSNRWTEMFP CIISRASTTD VISGGMAGTR 420 NGAIQLMHAE LQVLSPLVPV REANFLRFCK QHAEGVWAVV DVSVDGVREN SGCLPAMSLR 480 RLPSGCVVQD MPNGYSKVTW VEHAEYEENQ VHQLYRPMLR SGLGFGAQRW VATLQRQCEC 540 LAILMSSSVN PPDNGAITAG GRKSMLKLAQ RMTYNFCSGI SAPSVQSWSK LTVGHVGPDV 600 RVMTRKSVDD PGEPPGIVLS AATSVWVPGS PQRLFDFLRD ERLRCEWDIL SNGGPMQEMA 660 HIAKGQDEGN SVCLLRSNAM NASQSSMLIL QETRIDASGA LVVYAPVDIP AMHVVMNGGD 720 SSYVALLPSG FAILPDGGGP SSPHGGGGSP RGPASPHGGG VSPRPPAPRG SLLTVAFQIL 780 VNSLPTAKLT VESVETVNNL ITCTVQKIRA ALHSDS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
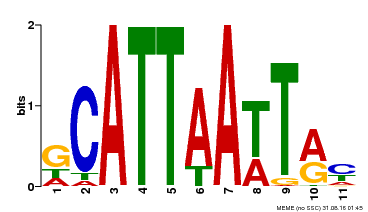
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010523042.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010523042.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A178V6U4 | 0.0 | A0A178V6U4_ARATH; ANL2 | ||||
| STRING | XP_010523042.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



