 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010523095.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 323aa MW: 35575.3 Da PI: 8.0277 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 30.4 | 7e-10 | 178 | 204 | 5 | 31 |
HHHHHHHHHHHHHHHHHHHHCTSCCC. CS
HLH 5 hnerErrRRdriNsafeeLrellPkas 31
hn+ Er+RRd+iN+++ +L++l+P++s
XP_010523095.1 178 HNQSERKRRDKINQRMKTLQKLVPNSS 204
9***********************973 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.44E-9 | 171 | 212 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 11.134 | 173 | 228 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.5E-10 | 177 | 223 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.7E-7 | 178 | 204 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.43E-7 | 178 | 202 | No hit | No description |
| SMART | SM00353 | 0.0036 | 179 | 229 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 323 aa Download sequence Send to blast |
MSQCVPTWDV DDSPTTARHS HRSHGAAADI PLLEEYEVAE LTWENGQLAL HGLGPPRAQT 60 QTKYVSGGTL ESIVNQATNS NNKGMDAMVP GSCRRDHPEE EAAAPKRPRR GDQISVSGSA 120 TVGRIRTTDS SGMGFTSTSL DDHYSFSPEM EVTHEEDTKN TAAKSSVSTK RSRAAAIHNQ 180 SERKRRDKIN QRMKTLQKLV PNSSKVSMMS RMSMPSMMLP MAMQQQLQMS MMANHMGLGM 240 GLGVMDMNSM NRAASNMMPN PFMPMAWDTA ASNDCRYPSP VIPDPMAAFL SCQSQPMTMD 300 AYSRMAALYQ QMQQHLPPPS NLK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 181 | 186 | ERKRRD |
| 2 | 182 | 187 | RKRRDK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Required during the fertilization of ovules by pollen. {ECO:0000269|PubMed:15634699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
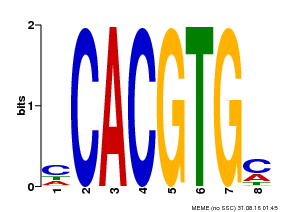
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010523095.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010523095.1 | 0.0 | PREDICTED: transcription factor UNE10 isoform X2 | ||||
| Swissprot | Q8GZ38 | 5e-84 | UNE10_ARATH; Transcription factor UNE10 | ||||
| TrEMBL | V4MYL2 | 4e-96 | V4MYL2_EUTSA; Uncharacterized protein | ||||
| STRING | XP_010523094.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 2e-52 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



