 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010523728.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 806aa MW: 88093 Da PI: 6.1197 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 63.9 | 2.2e-20 | 131 | 186 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F++ ++p++++r eL+++l+L+ rqVk+WFqNrR+++k
XP_010523728.1 131 KKRYHRHTPQQIQELESVFKEYPHPDEKQRLELSRRLNLDPRQVKFWFQNRRTQMK 186
688999***********************************************999 PP
| |||||||
| 2 | START | 185.8 | 2.2e-58 | 335 | 557 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....kaet 81
ela++a++elvk a+a+ep+Wv+ss e +n +e+ ++f++ v + +ea+r++g+v+ ++ lve+l+d+ +W e+++ + +t
XP_010523728.1 335 ELALAAMDELVKIAQAREPLWVRSSdsgrEVLNHEEYERNFSSC-VgpkregFVSEASREVGMVIINSLALVETLMDSE-RWAEMFPcmiaRTST 427
5899*********************8888888888888888654.36999999**************************.*************** PP
EEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE- CS
START 82 levissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwveh 169
+e issg galqlm+aelq+lsplvp R + f+R+++q+ +g+w++vdvS+d ++ + +++ +++lpSg+l+++++ng+skv wveh
XP_010523728.1 428 TEIISSGvggtrnGALQLMQAELQLLSPLVPvRRVSFLRFCKQHAEGVWAVVDVSIDTIRDSS--AGSPSCGRLPSGCLVQDMPNGYSKVKWVEH 520
*************************************************************76..56666777********************** PP
EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 170 vdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
++++++++h ++r+lv+sgla+gak+w a+lqrqce+
XP_010523728.1 521 AEYEESQIHRMYRPLVSSGLAFGAKRWIAALQRQCEC 557
***********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.68E-21 | 117 | 188 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-21 | 118 | 182 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.848 | 128 | 188 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.1E-19 | 129 | 192 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.00E-19 | 130 | 188 | No hit | No description |
| Pfam | PF00046 | 5.6E-18 | 131 | 186 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 163 | 186 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 40.531 | 326 | 560 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.88E-30 | 327 | 557 | No hit | No description |
| CDD | cd08875 | 4.56E-114 | 330 | 556 | No hit | No description |
| SMART | SM00234 | 1.2E-46 | 335 | 557 | IPR002913 | START domain |
| Pfam | PF01852 | 3.8E-50 | 335 | 557 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.33E-18 | 578 | 794 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 806 aa Download sequence Send to blast |
MDMHLSDKPL RFGFMNFSGF LDDSSGDAPR LLSDVSFANH FSSSAAAAIA QPHRGGFVPP 60 LAKPMFSSPG LSLGLQTNVE SNGEVTRGGE SFEVSANRRS REDEPESRSG SDNADGLSGD 120 DQDAAERPPR KKRYHRHTPQ QIQELESVFK EYPHPDEKQR LELSRRLNLD PRQVKFWFQN 180 RRTQMKTQLE RHENALLRQE NDRLRSENMS IREAVRNPIC GDCGGHAVLG EVSLEEQHLR 240 IENARLRDEL DRICALAGKF LGRPIPSHPL PDSALELGVG SNNAGFGVSS PPLPLVPPTG 300 FEIPAAGSGL APASRPPLAS TANVTGPDQR QMYLELALAA MDELVKIAQA REPLWVRSSD 360 SGREVLNHEE YERNFSSCVG PKREGFVSEA SREVGMVIIN SLALVETLMD SERWAEMFPC 420 MIARTSTTEI ISSGVGGTRN GALQLMQAEL QLLSPLVPVR RVSFLRFCKQ HAEGVWAVVD 480 VSIDTIRDSS AGSPSCGRLP SGCLVQDMPN GYSKVKWVEH AEYEESQIHR MYRPLVSSGL 540 AFGAKRWIAA LQRQCECLTI LMSSTVSSSF PTPISCSGRK SMLKLAQRMT ENFCGGVCAS 600 SAQKWSKLNV GNIDEDVRIM TRKSVDNPGE PPGIVLSAAT SVWLRGEGAR RLFDFLRDER 660 LRSEWDILSN GGPMQEMAHV AKGQDLSNCV SLLRASAMNA NQSSMLVLQE TCIDTAGALV 720 VYAPVDIPAM HVVMNGGDSA YVALLPSGFA IVPDSGQMEE GGSLLTVAFQ ILVNSLPTAK 780 LTVESVDTVN NLISCTVQKI RAALLP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00418 | DAP | Transfer from AT3G61150 | Download |
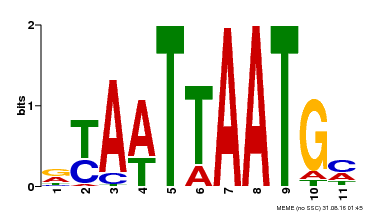
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010523728.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010523728.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG1-like | ||||
| Swissprot | Q9M2E8 | 0.0 | HDG1_ARATH; Homeobox-leucine zipper protein HDG1 | ||||
| TrEMBL | A0A067JXS1 | 0.0 | A0A067JXS1_JATCU; Uncharacterized protein | ||||
| STRING | XP_010523728.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



