 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010524575.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 923aa MW: 104090 Da PI: 6.935 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 163.4 | 3.9e-51 | 29 | 146 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+e k rwl+++ei+a+L n++ + + ++ + pksg+++L++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+nptf
XP_010524575.1 29 LEEaKGRWLRPNEIHAVLCNHKYFCINVKPMNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNEERIHVYYAHGEDNPTFV 123
455599***************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcywlL+++le+ivlvhy+e++
XP_010524575.1 124 RRCYWLLDKTLEHIVLVHYRETQ 146
********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 79.402 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 5.1E-76 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.2E-46 | 31 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.68E-11 | 369 | 453 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 4.94E-16 | 554 | 664 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.4E-17 | 555 | 667 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.95E-18 | 556 | 673 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.486 | 563 | 664 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.301 | 605 | 637 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2.0E-5 | 605 | 634 | IPR002110 | Ankyrin repeat |
| Pfam | PF13637 | 1.5E-5 | 607 | 652 | No hit | No description |
| SuperFamily | SSF52540 | 3.19E-7 | 714 | 819 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.181 | 753 | 779 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 230 | 768 | 790 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.002 | 791 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.871 | 792 | 816 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0015 | 794 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 11 | 869 | 891 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.334 | 870 | 899 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 923 aa Download sequence Send to blast |
MDGAGSGRLV GSDIHGFHTL QDLDVPTMLE EAKGRWLRPN EIHAVLCNHK YFCINVKPMN 60 LPKSGTIVLF DRKMLRNFRK DGHNWKKKKD GKTVKEAHEH LKVGNEERIH VYYAHGEDNP 120 TFVRRCYWLL DKTLEHIVLV HYRETQEVQT SPATPGNTSS ISDHSSPTCV AGDIDSGVGN 180 ARYVERNDPI AQNHEIRLHE INTLEWDELL VPNDDNSPPA PTVDDMSYLT QTLQNAANGS 240 AKNGNHLASY NASTDVQSFP CLGEPSYQNN NLCDPGRFSS QQVHCGVDPG LQNRDPSAAV 300 AGESVDALLN NGLQSQESFG RWMNAFISDS PGSVDDPSHE STLSPGQDSL TSPAAYHHQS 360 NMPEQIFSIT DVSPAWAFST EKTKILVTGF FHDGYQHQAR SNLFCICGET CVPAEIIQVG 420 VYRCFLPPQS PAIVNLYLSD DGQKPISQFF SFECRSAAVP EKSITQDNVS SRWEEFEFQV 480 RLAHLLFTSS NKVNILSSEV SAGSIQEAKK FVNKTSHLLN SWAYLVKSVQ GNQLSFEQAK 540 DNLFELTLKN RLKEWLLEKV LEGGKITEYD SKGLGVIHLC AILGYTWSIH LFSWSGLSLN 600 FRDKLGWTAL HWAAYYGREK MVAALLSAGA RPNLVTDPTK KNVDGCTAAD LAQQKGYDGL 660 AAYLSEKCLV AQFKDMKIAG NISGNLETCK AEASNPGTLT EEEQSLKDTL TAYRTAAEAA 720 SRIQVAFREH ALNVRSKAVQ FGSREEEAQS IIAAMKIQHA FRNHDTRKKM AAAVRIQYRF 780 HTWKMRREFL NMRRQVIKIQ AAFRGFQVRR QYRKIVWSVG VLEKAILRWR LKRKGFRGMQ 840 VSAITINEDE GETEEEDFYK TSQRQAEDRL ERSVVRVQAM FRSKKAQQDY RRMKLAHEEA 900 QLEYDGLQEL DDDDDDTSRA MER |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
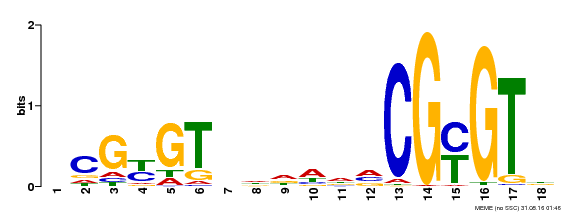
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010524575.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010524575.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A178UWV1 | 0.0 | A0A178UWV1_ARATH; Uncharacterized protein | ||||
| STRING | XP_010524574.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



