 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010524774.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 809aa MW: 90235.7 Da PI: 7.4715 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 117.2 | 8.7e-37 | 131 | 207 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cqv+gCead+se+k yhrrh+vC +++a++v+++g+++r+CqqC++fh ls+fDe+krsCrr+L++hn+rr++k
XP_010524774.1 131 RCQVPGCEADISELKGYHRRHRVCLRCANASSVVLDGESKRYCQQCGKFHVLSDFDEGKRSCRRKLERHNNRRKRKP 207
6************************************************************************9975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.4E-29 | 125 | 193 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.701 | 129 | 206 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.1E-34 | 130 | 209 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.4E-27 | 132 | 205 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 809 aa Download sequence Send to blast |
MSSASRSLPA QEMELQQPAM VDDDPSTFAA SIWEWGDLLD FDVDDRLLVS LDSDQPPAAA 60 APPPRHPTES ELYPSPGSGE DRVRKRDPRL TCSNFLEGRI PCSCPEIDQR LEDREFPTKK 120 RVRSGGRGVA RCQVPGCEAD ISELKGYHRR HRVCLRCANA SSVVLDGESK RYCQQCGKFH 180 VLSDFDEGKR SCRRKLERHN NRRKRKPVDN GGGSKKQQQA NMQNENSVID VDDGKDNICS 240 INQIAEQEAS LDFEDQHFSA QGAVPVVQSI NTDSFVSFAD QGGAQTVEGK NEKKIEHSPS 300 YGDNRSAYSS ECPTGRISFK LYDWNPAEFP RRLRHQIFQW LSTMPVELES YIRPGCTILT 360 VFIAMPEVMW AKLSKDPVAY LDEFILKPGK MLFGRGSMTV YLNNMIFGLM RGGNSVKSVN 420 VKIQAPRLQF VYPTCFEAGK PIELVICGRN LLQPKFRFLV SFCGKYLPYN YSVISPPNQD 480 GKSSPCCCNK LYKINVVNSD PNLFGPAFVE VENESGLSNF IPLIIGDEAI CSEMKLIEQK 540 LNATLFPDGQ VSACCSFMCF CGDFEQRQTA FSGLLLDIAW SVKVPISECT EQNMNLCQIR 600 RYNRVLDYLI QSNSASILAK VLQNMEILVN KMEPDSVIHC TSERDVRLLH ENMNLARDNF 660 RKCSSHEQST TNSGNIPPSS NRSCGCGCGW GCGSGCGCGC QSSFQKETPS RILTIINQDS 720 EAVRDSKQGM KPVVRKETDP LLSKEFVMNV DGVREWPVKS CSSASHAFRA RPTAFVVTTF 780 VVCLALCAVL YHPSKVTALA VAIRTRLAH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 5e-46 | 129 | 212 | 3 | 86 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
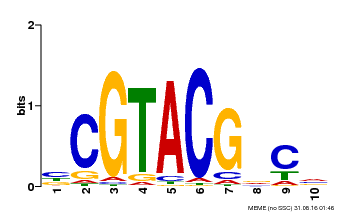
| UniProt | Transcription factor that participates in reprogramming global gene expression during copper deficiency in order to improve the metal uptake and prioritize its distribution to copper proteins of major importance (Probable). Binds directly to 5'-GTAC-3' motifs in the microRNA (miRNA) promoter of the stress-responsive miRNAs miR398b and miR398c to activate their transcription. During copper deficiency, activates the copper transporters COPT1 and COPT2, and the copper chaperone CCH, directly or indirectly via miRNAs. Required for the expression of the miRNAs miR397, miR408 and miR857 (PubMed:19122104). Acts coordinately with HY5 to regulate miR408 and its target genes in response to changes in light and copper conditions (PubMed:25516599). Activates miR857 and its target genes in response to low copper conditions (PubMed:26511915). Involved in cadmium stress response by regulating miR397a, miR398b, miR398c and miR857 (PubMed:27352843). Required for iron homeostasis during copper deficiency (PubMed:22374396). {ECO:0000269|PubMed:19122104, ECO:0000269|PubMed:22374396, ECO:0000269|PubMed:25516599, ECO:0000269|PubMed:26511915, ECO:0000269|PubMed:27352843}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010524774.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010524774.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 7 isoform X2 | ||||
| Swissprot | Q8S9G8 | 0.0 | SPL7_ARATH; Squamosa promoter-binding-like protein 7 | ||||
| TrEMBL | E4MYF5 | 0.0 | E4MYF5_EUTHA; mRNA, clone: RTFL01-52-E05 | ||||
| TrEMBL | V4N865 | 0.0 | V4N865_EUTSA; Uncharacterized protein | ||||
| STRING | XP_010524772.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.1 | 0.0 | squamosa promoter binding protein-like 7 | ||||



