 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010527982.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 246aa MW: 28173.1 Da PI: 7.9369 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 99.1 | 1.8e-31 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krienk+nrqvtf+kRrng+lKKA+ELSvLCdae+a+iifs++gklyey+s
XP_010527982.1 9 KRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEIALIIFSNRGKLYEYCS 59
79***********************************************96 PP
| |||||||
| 2 | K-box | 84.9 | 1.7e-28 | 82 | 174 | 8 | 100 |
K-box 8 sleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
++++++ ++ +q++ kL++++e Lq++qRhllGe++++L l +L+qLe q++ slk+iRs+K++++leqi+el++ke++l e+nk Lr klee
XP_010527982.1 82 NQSAKELQEKYQDFLKLRSRVEALQHSQRHLLGEGVGELGLDDLEQLELQVDMSLKQIRSTKTKFMLEQISELKRKEEMLMETNKGLRIKLEE 174
4688899*********************************************************************************99986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 2.9E-42 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 33.221 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 7.06E-33 | 2 | 79 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 3.24E-43 | 2 | 74 | No hit | No description |
| PRINTS | PR00404 | 4.7E-33 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 6.9E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.7E-33 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.7E-33 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.3E-24 | 85 | 172 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.789 | 88 | 178 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010076 | Biological Process | maintenance of floral meristem identity | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048441 | Biological Process | petal development | ||||
| GO:0048442 | Biological Process | sepal development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 246 aa Download sequence Send to blast |
MGRGKVELKR IENKINRQVT FAKRRNGLLK KAYELSVLCD AEIALIIFSN RGKLYEYCSS 60 PDGMANTLER YRQCRYETMD PNQSAKELQE KYQDFLKLRS RVEALQHSQR HLLGEGVGEL 120 GLDDLEQLEL QVDMSLKQIR STKTKFMLEQ ISELKRKEEM LMETNKGLRI KLEESEAAIN 180 QWQWGALEPG TSCPADPPNE REGSFIPSLQ SNGPLQISNY KPEVTNAATT SATTSQNVRV 240 FHGWMI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 2e-22 | 93 | 172 | 22 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 2e-22 | 93 | 172 | 22 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 2e-22 | 93 | 172 | 22 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 2e-22 | 93 | 172 | 22 | 101 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
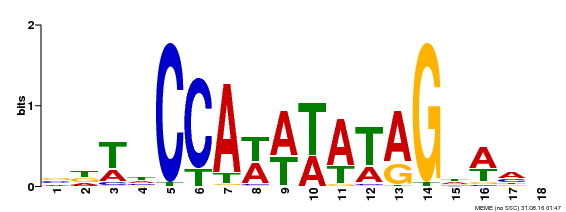
| UniProt | Probable transcription factor that binds specifically to the CArG box DNA sequence 5'-CC (A/T)6 GG-3' (PubMed:7632923, PubMed:25183521). Plays an important role in the determination of flower meristem identity. Involved in the specification of sepal identity. Contributes to the development of petals, stamens and carpels (PubMed:15530395). {ECO:0000269|PubMed:15530395, ECO:0000269|PubMed:25183521, ECO:0000269|PubMed:7632923}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00030 | SELEX | Transfer from AT2G03710 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010527982.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB623163 | 2e-40 | AB623163.2 Pyrus pyrifolia var. culta PpMADS9-1 mRNA for MADS-box protein, complete cds. | |||
| GenBank | AJ001681 | 2e-40 | AJ001681.1 Malus domestica mRNA for MADS box protein MdMADS8. | |||
| GenBank | KC415236 | 2e-40 | KC415236.1 Malus domestica cultivar Fuji Beni Shogun MADS9-2 mRNA, complete cds. | |||
| GenBank | KC415237 | 2e-40 | KC415237.1 Malus domestica cultivar Fuji Beni Shogun MADS8-2 mRNA, complete cds. | |||
| GenBank | KC415238 | 2e-40 | KC415238.1 Malus domestica cultivar Fuji Beni Shogun MADS8-1 mRNA, complete cds. | |||
| GenBank | KP164002 | 2e-40 | KP164002.1 Pyrus pyrifolia clone PpSEP1-1 SEP1-like MADS-box protein mRNA, complete cds. | |||
| GenBank | KP164016 | 2e-40 | KP164016.1 Pyrus pyrifolia clone PpSEP1-2 SEP1-like MADS-box protein mRNA, complete cds. | |||
| GenBank | U78947 | 2e-40 | U78947.1 Malus domestica MADS-box protein 1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010527982.1 | 0.0 | PREDICTED: agamous-like MADS-box protein AGL3 | ||||
| Swissprot | P29383 | 1e-111 | AGL3_ARATH; Agamous-like MADS-box protein AGL3 | ||||
| TrEMBL | A0A0D3DAN8 | 1e-113 | A0A0D3DAN8_BRAOL; Uncharacterized protein | ||||
| STRING | XP_010527982.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7135 | 24 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03710.1 | 2e-96 | MIKC_MADS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



