 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010528431.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 274aa MW: 30812.9 Da PI: 6.5221 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40.5 | 5.1e-13 | 38 | 93 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHH......HHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiL......ekAveYIksLq 55
r +h+e E+rRR++iN++f++L +l+P++ ++K +Ka+ L +++eYI+ Lq
XP_010528431.1 38 RSKHSETEQRRRSKINERFQTLMDLIPQN----DQKRDKASFLlelfyvSQVIEYIHFLQ 93
899*************************9....89******9844444457999999888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 5.4E-21 | 34 | 113 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.8E-16 | 35 | 118 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 12.551 | 36 | 92 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 9.0E-11 | 38 | 93 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.37E-13 | 38 | 97 | No hit | No description |
| SMART | SM00353 | 2.0E-7 | 42 | 98 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 274 aa Download sequence Send to blast |
MKSPDLEEQE DNDVYDNLPC RNGSSSGEVT RRKTSTYRSK HSETEQRRRS KINERFQTLM 60 DLIPQNDQKR DKASFLLELF YVSQVIEYIH FLQEKLQMYE GSHQMWNQGS AKLIPWRNNP 120 RPAEDASDHS QGMKSWTLNQ DPTSELPSYF PAHPTLQFVQ QDSCQVRPSS GSAVCNTSEP 180 SNLKEKNLAS SSTAYSQGVL SSLTEALKSS GVNLSETLIS VQLSVGKRAD PEAAVTASAS 240 AENTNGLADR EVGSQGETRS FYEESKHAQK RIRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00534 | DAP | Transfer from AT5G38860 | Download |
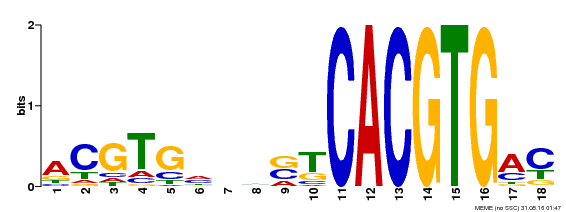
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010528431.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010528431.1 | 0.0 | PREDICTED: transcription factor BIM3 isoform X2 | ||||
| Swissprot | Q9FMB6 | 6e-93 | BIM3_ARATH; Transcription factor BIM3 | ||||
| TrEMBL | D7MJT9 | 2e-92 | D7MJT9_ARALL; Uncharacterized protein | ||||
| STRING | XP_010528430.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G38860.1 | 3e-95 | BES1-interacting Myc-like protein 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



