 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010529031.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 360aa MW: 40881.2 Da PI: 6.6868 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.6 | 5.1e-32 | 67 | 120 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH rFv+av++LGG+e+AtPk +l+lm++kgL+++hvkSHLQ+YR+
XP_010529031.1 67 PRLRWTPDLHLRFVHAVDRLGGQERATPKLVLQLMNIKGLSIAHVKSHLQMYRS 120
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.593 | 63 | 123 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-29 | 63 | 121 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 8.42E-15 | 65 | 122 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-22 | 67 | 122 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.2E-9 | 68 | 119 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 360 aa Download sequence Send to blast |
MEGGGDAEYS KTSRYERIEI NESGEEEEEE EEEEEGGGGE SKASSNSTVE ESDKKPKVRP 60 YVRSKVPRLR WTPDLHLRFV HAVDRLGGQE RATPKLVLQL MNIKGLSIAH VKSHLQMYRS 120 KKTDDQCQAM AEHRHLIEST TDRNIYNLSQ LPMFRAYSHD HDSPFRYGNA SMWNSSSHVP 180 DRSLIDQLRP GFVRGSVVNN WANNMMNRSF QNISSSLVSS HLGMLRQNRT QILDQDSPGT 240 FKNIQDQRSG TINKLELHHD IAQETDNNAY YNKMGKRKAS ERIDLDLDLS LKLARHEKAS 300 LDETERDQTL SLSLGSGSSS WKQSRTLKKT DEEDDNGTAK RLLSSHEHGI GRASTLDLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 4e-17 | 68 | 122 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-17 | 68 | 122 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-17 | 68 | 122 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-17 | 68 | 122 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
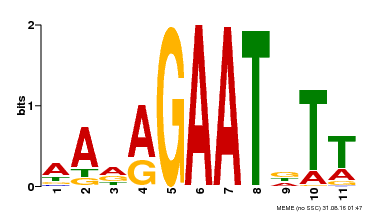
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010529031.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010529031.1 | 0.0 | PREDICTED: uncharacterized protein LOC104806008 | ||||
| TrEMBL | A0A178VYP6 | 1e-116 | A0A178VYP6_ARATH; Uncharacterized protein | ||||
| TrEMBL | Q8GYE4 | 1e-116 | Q8GYE4_ARATH; Myb-like HTH transcriptional regulator family protein | ||||
| STRING | XP_010529031.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM363 | 28 | 184 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 1e-113 | G2-like family protein | ||||



