 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010529274.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 673aa MW: 75321.3 Da PI: 6.3141 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 158.5 | 4.4e-49 | 70 | 209 | 1 | 136 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
g++g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+V++AL+reAGw+ve DGttyr++ p+ ++ +++++ +s+es+l+
XP_010529274.1 70 GAKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVIAALAREAGWTVETDGTTYRQS--PP--TNPVVHFQARSVESPLS 160
5899******************************************************************55..33..35666677777777776 PP
DUF822 96 .sslkssalaspvesysaspksssfpspssldsislasa.......asl 136
++lks+++++ ++ ++++++ ++ +sp+slds+ +a++ +s
XP_010529274.1 161 sNTLKSCSVKAALDCQQSVLRIDENLSPVSLDSVVIAESdagneryTSA 209
678*********************************9886665444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.4E-48 | 72 | 215 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 7.62E-161 | 236 | 671 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 3.1E-172 | 239 | 670 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.2E-85 | 247 | 633 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 276 | 290 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 297 | 315 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 319 | 340 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 412 | 434 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 485 | 504 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 519 | 535 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 536 | 547 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 554 | 577 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.8E-57 | 592 | 614 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 673 aa Download sequence Send to blast |
MHTLNNTTTD SQDPNFDPDQ VSYRTQQQQP LSQPRRPRGF AAAAAAGGAA GSDNGDSGSI 60 RGGGGSGGGG AKGKREREKE KERTKLRERH RRAITSRMLA GLRQYGNFPL PARADMNDVI 120 AALAREAGWT VETDGTTYRQ SPPTNPVVHF QARSVESPLS SNTLKSCSVK AALDCQQSVL 180 RIDENLSPVS LDSVVIAESD AGNERYTSAS PINSVGGLEA DQLIQDVRSA QHQNAFTGNC 240 YVPVYTMLPA GIINNFCQLV DPEAVRQELS YMKSLNVDGV VIECWWGIVE GWNPKKYVWS 300 GYRELFNIVR DFKLKLQVVM AFHEYGGNPS SNAIISLPQW VLEIAKDNPD IFFTDREGRR 360 NTECLNWSID KERVLHGRTG IEVYFDVMRS FRTEFDDLFA EGLISAVEIG LGASGELKYP 420 SFSERMGWSY PGIGEFQCYD KYSQKNLQKA AKSREYSFWA KGPENAGHYN SRPHETGFFC 480 DRGDYDSYYG RFFLNWYSQL LIEHAENVLS LASLAFDETK IIVKVPAIYW WYKTASHAAE 540 LTAGYYNPTN RDGYSPVFEA LKKHSVTVKF VCSGLQMLTT EHDKALADPE GLSWQVLNAA 600 WDGGMSIAGE NAVSCFDREG CMRIVEMAKP RNHPENRHFS FFVYHQPSPL VQGSTCFADL 660 DYFIKCMHGD IQR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-119 | 241 | 670 | 11 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
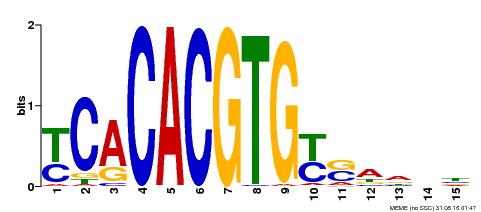
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010529274.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010529274.2 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | D7MUR1 | 0.0 | D7MUR1_ARALL; Beta-amylase | ||||
| STRING | XP_010529274.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10888 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



