 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010529979.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 261aa MW: 29307 Da PI: 8.7509 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 62.5 | 6.3e-20 | 117 | 171 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ +++k+q +Le+ F+++++++ +++ LAk+lgLt rqV vWFqNrRa+ k
XP_010529979.1 117 RKKLRLSKDQSAILEDTFKEHNTLNPKQKMALAKQLGLTPRQVEVWFQNRRARTK 171
788899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.4 | 3e-41 | 117 | 206 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
+kk+rlsk+q+++LE++F+e+++L+p++K++la++Lgl+prqv+vWFqnrRARtk+kq+E+d+e+Lkr++++l+e+n+rL+kev+eLr +l
XP_010529979.1 117 RKKLRLSKDQSAILEDTFKEHNTLNPKQKMALAKQLGLTPRQVEVWFQNRRARTKLKQTEVDCEYLKRCVEKLTEDNRRLQKEVAELR-AL 206
69*************************************************************************************9.55 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04618 | 5.0E-29 | 4 | 99 | IPR006712 | HD-ZIP protein, N-terminal |
| SuperFamily | SSF46689 | 9.19E-20 | 106 | 174 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.734 | 113 | 173 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.6E-18 | 115 | 177 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 3.2E-17 | 117 | 171 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-19 | 117 | 171 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 4.52E-17 | 117 | 174 | No hit | No description |
| PRINTS | PR00031 | 3.5E-6 | 144 | 153 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 148 | 171 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 3.5E-6 | 153 | 169 | IPR000047 | Helix-turn-helix motif |
| SMART | SM00340 | 6.4E-25 | 173 | 216 | IPR003106 | Leucine zipper, homeobox-associated |
| Pfam | PF02183 | 2.0E-9 | 173 | 207 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0048467 | Biological Process | gynoecium development | ||||
| GO:0080127 | Biological Process | fruit septum development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0043621 | Molecular Function | protein self-association | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 261 aa Download sequence Send to blast |
MMMMTEEEDM GLSLSLGFAQ NHRPLQLNLK PTSFVSNLHK FPWNQTFVSS AADHQNPPFL 60 GGIDVNRAPS TGDCEEEAGV SSPNSTISST ISGKRSEREG NDHRVSSDEE DGGETSRKKL 120 RLSKDQSAIL EDTFKEHNTL NPKQKMALAK QLGLTPRQVE VWFQNRRART KLKQTEVDCE 180 YLKRCVEKLT EDNRRLQKEV AELRALKLSP QFYGQMAPPT TLIMCPSCER VAGSSNQTAQ 240 RDRSRPLNPW LSGPSRMAKG S |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 115 | 121 | SRKKLRL |
| 2 | 165 | 173 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00050 | PBM | Transfer from AT4G17460 | Download |
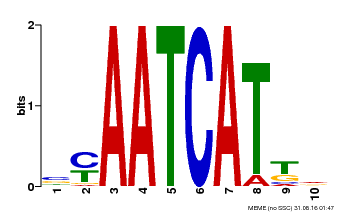
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010529979.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010529979.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HAT1 | ||||
| Swissprot | P46600 | 1e-126 | HAT1_ARATH; Homeobox-leucine zipper protein HAT1 | ||||
| TrEMBL | A0A1J3FTB7 | 1e-128 | A0A1J3FTB7_NOCCA; Homeobox-leucine zipper protein HAT1 | ||||
| STRING | XP_010529979.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1563 | 26 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17460.1 | 1e-119 | Homeobox-leucine zipper protein 4 (HB-4) / HD-ZIP protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



