 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010537252.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1058aa MW: 117859 Da PI: 8.6604 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 127.3 | 5.8e-40 | 146 | 223 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv+ C++dls+ak+yhrrhkvC+ hska+++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+q+
XP_010537252.1 146 MCQVDFCKEDLSNAKDYHRRHKVCKLHSKASKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLAGHNRRRRKTQP 223
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.0E-33 | 141 | 208 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.547 | 144 | 221 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 7.06E-37 | 145 | 226 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.4E-28 | 147 | 220 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 3.1E-4 | 843 | 978 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.53E-5 | 854 | 980 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1058 aa Download sequence Send to blast |
MDEVGAQVAA PVFVHQSLTP MGRKRDLYFQ IPNLHTHRLP QTQTLQRGDD WNPKMWDWDS 60 RRFEAKPVET EVLRLGNDSQ FDVSAKSNNN NSGLTLMNSG CGGSEVDERN LDLNLGSGLT 120 GMEETQANRQ NKRVRSGSPG GGNYPMCQVD FCKEDLSNAK DYHRRHKVCK LHSKASKALV 180 GKQMQRFCQQ CSRFHLLSEF DEGKRSCRRR LAGHNRRRRK TQPEEITSRV VVPGNRDPNS 240 NPNTDLVALL TALACAQGRN EVKPIESTVV PDRERLLQIL NKINSLPLPM DLASKLNNIG 300 VLARKNLDHS VDNFRNDMNG TSQSTMDLLA VLSSTLASSS HDALALMSQM PVEISDSDKT 360 KLTPSDQGTT ADFEKRTRDF GSVGGEKSSS SHQSPSQDSD SHFQDTRSSL SLQLFTSSPE 420 DESRPTVATS RKYYSSASSN PVEDRSLSSS PVMQELFPKQ ASPETTRSKK DKNTGFRTGC 480 LPLELFGASN RGAENNIFKG FGHQAGYASS GSDNSPPSLN SDAQDRTGRI VFKLLGKDPS 540 QLPGTLRTQI YNWLSNIPSE MESYIRPGCV ILSVYVAMSS AAWEQLEENL QERVGFLLQD 600 SCSEFWRRTR FLVNTGRQLA SYKDGKIRLS KSWRTWNSPE LISVSPVAVV AGEETTLLVR 660 GRNLDNGEIR IRCAHMGSYA SMEVTTSSCK KPVFDELNVS SFRINGGSHG FFGRCFIEVE 720 NGFRGDSFPL IIADAAICKE LNRLEDEFHP KTQVHRDITE ERLHNSDRPS SREEVLCFLN 780 ELGWLFQRNA SSFDIQGGSD FSMTRFKFLL VFSVERDYCS LVRKLLDMVV ERNSGKDGLI 840 RESRDMLAEI QLLNRAVKRR STKMAKMLIH YSVTASSLST SKEFVFLPDT AGPGGITPLH 900 LAASTSGSDG MIDVLTNDPQ EIGLSCWNSL LDKSGHTPYS YASTRNNHRY NALVARKLTD 960 KRNRQVSINI ESRIDGQTGL SNRSSSELKR SSCTTCGTVA LKYQRKISGS YRLFPTPIIH 1020 SMLAVATVCV CVCVFMHAFP IVRQGAHFSW GGLDYGSM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 7e-30 | 137 | 220 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 854 | 859 | RAVKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
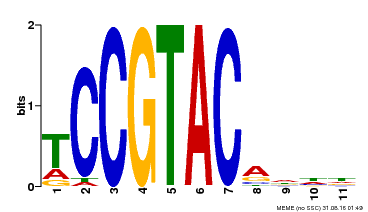
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010537252.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010537252.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | V4KAJ1 | 0.0 | V4KAJ1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_010537252.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



