 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010537716.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 309aa MW: 33495.2 Da PI: 8.4605 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 183.4 | 9.9e-57 | 2 | 133 | 1 | 149 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
++++r+ptw+ErEnnkrRERrRRa+aaki+aGLR++Gnyklpk++DnneVlkALc+eAGw+ve+DGttyrkg+kp + + asp+ss q
XP_010537716.1 2 TSGTRMPTWRERENNKRRERRRRAMAAKIFAGLRMYGNYKLPKHCDNNEVLKALCNEAGWIVEPDGTTYRKGCKPV------ERTNYASPCSSFQ 90
5899************************************************************************......4555677777777 PP
DUF822 96 sslkssalaspvesysaspksssfpspssldsislasaasllpvlsvlslvsss 149
+sp +sys sp+s s+ ++l + a+ +sl+p+l++ls++sss
XP_010537716.1 91 --------PSPFTSYSHSPASASSSLAANLTT---ADGQSLIPWLRNLSTSSSS 133
........777777777777665544444433...3446777777777765444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.0E-51 | 3 | 128 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 309 aa Download sequence Send to blast |
MTSGTRMPTW RERENNKRRE RRRRAMAAKI FAGLRMYGNY KLPKHCDNNE VLKALCNEAG 60 WIVEPDGTTY RKGCKPVERT NYASPCSSFQ PSPFTSYSHS PASASSSLAA NLTTADGQSL 120 IPWLRNLSTS SSSSSSSRFP YLYVPGGSIS APVTPPLSSP TARTPRMKPD FQQNNSFFFY 180 STPPSPTRQQ LVPDPEWFSG IQIPQSVPTS PTFSLVSANP FGFKEASGNN GSGGSMMWTP 240 GQSGTCSPAA IPPGSDQTAD VPMSEAVPAE FAFGGNENGL VKAWEGERIH EESGSDDLEL 300 TLGNSTTRQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 9e-24 | 6 | 77 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 9e-24 | 6 | 77 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 9e-24 | 6 | 77 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 9e-24 | 6 | 77 | 372 | 443 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
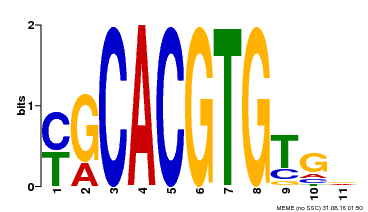
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010537716.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010537716.1 | 0.0 | PREDICTED: BES1/BZR1 homolog protein 4 | ||||
| Swissprot | Q9ZV88 | 1e-144 | BEH4_ARATH; BES1/BZR1 homolog protein 4 | ||||
| TrEMBL | A0A178WLH5 | 1e-142 | A0A178WLH5_ARATH; BEH4 | ||||
| STRING | XP_010537716.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1917 | 27 | 81 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 1e-138 | BES1/BZR1 homolog 4 | ||||



