 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010538501.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 646aa MW: 69885 Da PI: 5.0546 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 36.2 | 1.1e-11 | 470 | 515 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ A+ YI++L
XP_010538501.1 470 NHVEAERQRREKLNQRFYALRAVVPNV-----SKMDKASLIGDAISYISEL 515
799***********************6.....5****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 8.8E-53 | 71 | 257 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.806 | 466 | 515 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 8.77E-18 | 469 | 533 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.82E-14 | 469 | 520 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.8E-17 | 470 | 535 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.3E-9 | 470 | 515 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.2E-15 | 472 | 521 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 646 aa Download sequence Send to blast |
MADYRQNQPT KGTTASINQW TTDDDASVME AFMGSDHSAL WPPATPHQQL SGGAHPQQPA 60 AAQSQFNEDT LQQRLQSLIE GAREPWTYAI FWQLSYDFPG GDGTALLGWG DGYYKGEEDK 120 GKSRKKCSSA SAAEQEHRKR VIRELNALIS GGEAVGAADD AGDEEVTDTE GFFLVSMTQS 180 FLNGTGLPGQ ASLNSRTIWL AGSGVLAGSG CERARQGQIY GLQTLVCIPS ENGVVELGST 240 EVIQQSSDLM EKVNTLFHFN DGGGEAGSWA FSLNPDEGEN DPALWISEPT TGIESGHVAP 300 TEAINNGNSN SNSNSNSHLF PKPGHFDNGG SSVENPGQNP QSDLNLSSSG LNQNGNYPDG 360 SRPKSGEILT SSEKRNGNGT GYSGQTSFMA GDSNKKRSPV SKGSNNEEGM LSFTCGVVIP 420 SAAAKSGDSD HSDVEASVAK EADSGRTVEP EKRPRKRGRK PANGREEPLN HVEAERQRRE 480 KLNQRFYALR AVVPNVSKMD KASLIGDAIS YISELKSKLQ TAESDKEELR QQLNKEESGK 540 DSRPQQRSKD RKGVITNQDS GLAIEMEIDV KIIGWDVMIR IQCGKKNHPG AKFMEALKDL 600 DLEVNHASLS VVNDLMIQQA TVKMGNRFFS QDQLKRALMA KVGEIQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rru_A | 2e-91 | 14 | 253 | 1 | 227 | Transcription factor MYC3 |
| 4ywc_A | 2e-91 | 14 | 253 | 1 | 227 | Transcription factor MYC3 |
| 4ywc_B | 2e-91 | 14 | 253 | 1 | 227 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 451 | 459 | KRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in jasmonic acid (JA) gene regulation. With MYC2 and MYC3, controls additively subsets of JA-dependent responses. Can form complexes with all known glucosinolate-related MYBs to regulate glucosinolate biosynthesis. Binds to the G-box (5'-CACGTG-3') of promoters. Activates multiple TIFY/JAZ promoters. {ECO:0000269|PubMed:21321051, ECO:0000269|PubMed:21335373, ECO:0000269|PubMed:23943862}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00086 | PBM | Transfer from AT4G17880 | Download |
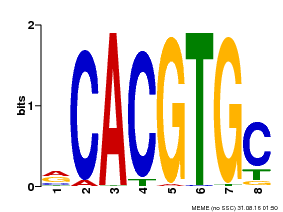
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010538501.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. Not induced by jasmonic acid. {ECO:0000269|PubMed:12679534, ECO:0000269|PubMed:21335373}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010538501.1 | 0.0 | PREDICTED: transcription factor MYC4-like | ||||
| Swissprot | O49687 | 0.0 | MYC4_ARATH; Transcription factor MYC4 | ||||
| TrEMBL | E4MW10 | 0.0 | E4MW10_EUTHA; mRNA, clone: RTFL01-05-E20 | ||||
| TrEMBL | V4L837 | 0.0 | V4L837_EUTSA; Uncharacterized protein | ||||
| STRING | XP_010538501.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1265 | 28 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17880.1 | 0.0 | bHLH family protein | ||||



