 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_010542803.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Cleomaceae; Tarenaya
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 747aa MW: 81776.4 Da PI: 6.607 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 47.9 | 3.2e-15 | 24 | 68 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT++E+ ++++a +++G W +I +++g ++t+ q++s+ qk+
XP_010542803.1 24 RERWTEDEHSRFLEALRLYGRA-WQRIEEHIG-TKTAVQIRSHAQKF 68
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.39E-15 | 18 | 72 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.323 | 19 | 73 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-7 | 21 | 70 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.1E-17 | 22 | 72 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.0E-11 | 23 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.8E-12 | 24 | 67 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.54E-8 | 26 | 69 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 747 aa Download sequence Send to blast |
MEKNISGEEL VTKIRKPYTI TKQRERWTED EHSRFLEALR LYGRAWQRIE EHIGTKTAVQ 60 IRSHAQKFFT KQPYAWKAST WMRFAVDASI ELHKKLLEKE AQAKGIPICQ ALDIEIPPPR 120 PKRKPSNPYP RKTGNGESTS QVSSAKVVKL DSSASFSHCN QALDLEKKPF TEPNGVEKVP 180 NGNGNQDDNC SGVSARPTEA HCSSISSVNK YPFPNKAAPR NPSSFRESEP DLTKATADNG 240 TSKTSNADNS CYSQDNDANV LPHISISKED NGVNGLSPVG NMNGIQSYPW HFPVHVLNGN 300 VGTYPQNPPG MGFQDFMFRP VGEVHGHSGL FTIPSTSATS QHQDNVPRSS RQAFPAFHPA 360 FVPACHTQDD YRSFLQISST FSNLIMSALL QNPAAHAAAT FAASFWPYGN VGSSNDSSTS 420 TTIQTDGAFP PRQMSSPPPS MAAITAATVA AATAWWAAHG LLPICAPFTC LPMSAPIVPP 480 SVSAQTELPE TDKKDAATAN PQQPAMEQGE AQVLASPFES DDDLDENGRT EQDTGLKTED 540 HKEKVVEVQE QVQDSNTTQK KKQVDRSSCG SNTPSGSDVE TDALEKAAEK DKEEDVKETD 600 GNQTGTESST RRTSRSKDNS SDSWKEVSEE GRMAFDALFA RERLPQSFSL ALDVEKESPE 660 KGKGNREERE REMAHKSKIQ DSCSKETASR SEGNGGITIG IGEGKSLRAR RTGFKPYKRC 720 SMEAKESSSQ AEEKVSKRIR LDSKAST |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the circadian clock. Binds to the promoter region of APRR1/TOC1 and TCP21/CHE to repress their transcription. Represses both CCA1 and itself. {ECO:0000269|PubMed:12015970, ECO:0000269|PubMed:19095940, ECO:0000269|PubMed:19218364, ECO:0000269|PubMed:9657154}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00654 | PBM | Transfer from LOC_Os08g06110 | Download |
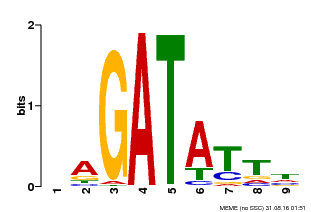
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_010542803.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation with peak levels occurring around 1 hour after dawn. Up-regulated by APRR1/TOC1 and transiently by light treatment. Down-regulated by APRR5, APRR7 and APRR9. {ECO:0000269|PubMed:12574129, ECO:0000269|PubMed:19095940, ECO:0000269|PubMed:19218364, ECO:0000269|PubMed:19286557, ECO:0000269|PubMed:20233950, ECO:0000269|PubMed:9657154}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010542803.1 | 0.0 | PREDICTED: protein LHY-like isoform X2 | ||||
| Swissprot | Q6R0H1 | 0.0 | LHY_ARATH; Protein LHY | ||||
| TrEMBL | A0A061DT31 | 0.0 | A0A061DT31_THECC; Homeodomain-like superfamily protein isoform 1 | ||||
| STRING | XP_010542800.1 | 0.0 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01060.4 | 1e-144 | MYB_related family protein | ||||



